doi: 10.56294/dm2023112
ORIGINAL
Enhancing Plant Disease Classification through Manual CNN Hyperparameter Tuning
Mejora de la clasificación de enfermedades vegetales mediante el ajuste manual de hiperparámetros de CNN
Khaoula Taji1 *, Fadoua Ghanimi2 *
1Electronic Systems, Information Processing, Mechanics and Energy laboratory, Ibn Tofail University. Moroco.
2Faculty of Sciences. Kenitra, Morocco.
Cite as: Taji K, Ghanimi F. Enhancing Plant Disease Classification through Manual CNN Hyperparameter Tuning. Data and Metadata. 2023;2:112. https://doi.org/10.56294/dm2023112
Submitted: 21-08-2023 Revised: 20-10-2023 Accepted: 26-12-2023 Published: 27-12-2023
Editor: Prof.
Dr. Javier González Argote ![]()
ABSTRACT
Diagnosing plant diseases is a challenging task due to the complex nature of plants and the visual similarities among different species. Timely identification and classification of these diseases are crucial to prevent their spread in crops. Convolutional Neural Networks (CNN) have emerged as an advanced technology for image identification in this domain. This study explores deep neural networks and machine learning techniques to diagnose plant diseases using images of affected plants, with a specific emphasis on developing a CNN model and highlighting the importance of hyperparameters for precise results. The research involves processes such as image preprocessing, feature extraction, and classification, along with a manual exploration of diverse hyperparameter settings to evaluate the performance of the proposed CNN model trained on an openly accessible dataset. The study compares customized CNN models for the classification of plant diseases, demonstrating the feasibility of disease classification and automatic identification through machine learning-based approaches. It specifically presents a CNN model and traditional machine learning methodologies for categorizing diseases in apple and maize leaves, utilizing a dataset comprising 7023 images divided into 8 categories. The evaluation criteria indicate that the CNN achieves an impressive accuracy of approximately 98,02 %.
Keywords: Plant Disease; Disease Classification; Machine Learning (ML); Convolutional Neural Network (CNN); Hyper-parameters; Plant Disease; Plant Disease Detection; Classification.
RESUMEN
El diagnóstico de las enfermedades de las plantas es una tarea difícil debido a la naturaleza compleja de las plantas y a las similitudes visuales entre las distintas especies. La identificación y clasificación oportunas de estas enfermedades son cruciales para evitar su propagación en los cultivos. Las redes neuronales convolucionales (CNN) han surgido como una tecnología avanzada para la identificación de imágenes en este ámbito. Este estudio explora las redes neuronales profundas y las técnicas de aprendizaje automático para diagnosticar enfermedades vegetales utilizando imágenes de plantas afectadas, con un énfasis específico en el desarrollo de un modelo CNN y destacando la importancia de los hiperparámetros para obtener resultados precisos. La investigación incluye procesos como el preprocesamiento de imágenes, la extracción de características y la clasificación, junto con una exploración manual de diversos ajustes de hiperparámetros para evaluarel rendimiento del modelo CNN propuesto entrenado en un conjunto de datos de libre acceso. El estudio compara modelos CNN personalizados para la clasificación de enfermedades de plantas, demostrando la viabilidad de la clasificación e identificación automática de enfermedades mediante enfoques basados en el aprendizaje automático. En concreto, presenta un modelo CNN y metodologías tradicionales de aprendizaje automático para clasificar enfermedades en hojas de manzana y maíz, utilizando un conjunto de datos compuesto por 7 023 imágenes divididas en 8 categorías. Los criterios de evaluación indican que la CNN alcanza una impresionante precisión de aproximadamente el 98,02 %.
Palabras clave: Enfermedad vegetal; Clasificación de Enfermedades; Aprendizaje Automático (ML); Red Neuronal Convolucional (CNN); Hiperparámetros; Enfermedad Vegetal, Detección de Enfermedades Vegetales; Clasificación.
INTRODUCTION
Plant diseases can significantly reduce production and quality, which can have a disastrous effect on agricultural output. For these losses to be reduced and sustainable agricultural output to be achieved, it is essential to be able to identify and categorize plant lesions properly and effectively.(1)Various methods, from conventional ones like eye inspection to cutting-edge ones like machine learning and deep learning,(2) have been developed in recent years to automate the identification and categorization of plant diseases.(3) These methods might significantly improve the efficiency and efficacy of disease control measures, empowering farmers and agricultural workers to quickly identify and address plant illnesses. The identification and categorization of plant diseases can have broader environmental and social advantages in addition to increasing crop quality and output.(4) Farmers may support more sustainable and eco-friendly farming practices by using fewer pesticides and other dangerous chemicals. Additionally, improved disease management techniques(5) can contribute to better food security and economic development, especially in areas where agriculture is a major source of income.(6) On the basis of images of diseased plants, machine learning, and deep learning have demonstrated promising results in properly categorizing and detecting plant diseases.(7) The development and application of image recognition techniques for spotting leaf diseases is currently one of the agricultural research's most passionately debated topics. Crop infections can inflict major harm in the field and are challenging to identify over vast regions of devoted agriculture because crop disease categorization rests on the expert's naked eye observations. There are several agricultural uses for artificial intelligence (AI).(8) The quantity of land needed to feed the growing population decreases as cities develop. Plant diseases have a severe negative influence on crop yield, which causes agriculture to suffer huge financial losses. They may harm the fruits as well as the leaves, stems, and roots.(9) Agriculture diseases are the primary cause of food shortages and rising food production costs, accounting for an estimated 16 % of global crop output loss each year.(10) In order to fulfill the demands of a growing population, the FAO study emphasizes the crucial significance of expanding food production. This is especially important in poorer nations where obtaining enough food is already a huge challenge. It will take a lot of work and money to meet this need in agricultural research, development, and innovation. Precision farming and crop management strategies are two sustainable agricultural practices that can assist in boosting yields while reducing environmental consequences. Additionally, advances in biotechnology and genetic engineering have led to the development of crop varieties that use less water, are more resistant to pests and diseases, and can endure in harsher climates. The employment of these technologies must be carefully weighed against any possible ethical, social, and environmental ramifications.(11) The factors that affect plants and their products are referred to as diseases and disorders, respectively. Algae, fungi, and bacteria are biotic factors that cause disease, whereas rainfall, humidity, temperature, and nutritional deficiency are biotic factors that cause disorders.(12) The factors that affect plants and the goods they produce are referred to as diseases and disorders, respectively. While rainfall, moisture, temperature, and nutritional insufficiency are some of the biotic variables that induce problems, algae, fungi, and bacteria are the biotic factors that cause illnesses.(13) It is essential to identify plant illnesses for accurate diagnosis, and efficient treatment, and to halt the spread of plant diseases. It is necessary for analyzing plant diseases(14) and creating cutting-edge preventative measures. One of the most common and straightforward techniques for diagnosing plant diseases(15) is visual estimate. The traditional approaches to identifying plant diseases rely on the farmer's expertise, which is inherently unreliable. Instead of using standard plant disease detection methods, the researchers used a spectrometer to determine if plant leaves were healthy or diseased.(16) Another method is to take the DNA from the leaves using real-time polymerase chain reaction or polymerase chain reaction.(17) Such techniques demand highly skilled operation, substantial testing, and frequent application of crop protection chemicals. They are also complicated, expensive, and time-consuming. Automation of plant leaf disease detection methods has been made possible by recent advancements in machine learning, artificial intelligence (AI), and computer vision (CV) technology.(18) Without involving humans, these techniques may accurately and swiftly identify plant leaf disease. In recent decades, a number of methods for identifying leaf disease in various crops have been developed.(19) By differentiating between sick and healthy plants, an intelligent classification system that allows for the targeted application of fungicides may assist in improving economic and environmental sustainability. In this work, we compare models that employ machine-learning techniques to detect early-stage infections of various leaf diseases, such as leaf blight, leaf spot, and leaf rust. RF, LR, XGboost, DT, NB, SVM, and CNN are examples of supervised algorithms for machine learning that may be used to classify illnesses from plant leaves. The rest of this essay is structured as follows. In Section 2, related works are discussed. The proposed method is covered in full in section 3. With the aid of the findings of the experiments, Section 4 describes the effectiveness of the suggested technique. Finally, Section 5 has provided the conclusion and suggestions for more research.
Related work
Plant diseases pose a serious danger to agricultural production and global food security, resulting in large financial losses and food shortages. Effective management and control of the spread of plant diseases depend on the early diagnosis and precise categorization of plant diseases. Convolutional neural networks (CNNs), in particular, have recently demonstrated considerable promise in reliably diagnosing and categorizing plant illnesses based on photographs of sick plants. For classifying plant diseases, a variety of CNN models have been put forth, each with unique advantages and disadvantages. To determine the most effective and efficient method for classifying plant diseases, it is crucial to compare and analyze different models. A deep convolutional neural network for classifying Potato Leaf illnesses was suggested by the authors in.(9) Two major convolutional layers for feature extraction are followed by two fully connected layers for classification in the architecture, which has 14 layers altogether. They utilized a data augmentation technique that raised the number of photos in the data set from 1 722 to 9 822. As a result, the model's performance was enhanced, going from 94,8 % to 98 % accuracy. They suggested a detection and classification method in,(20) to address the problem of Potato Leaf Diseases. K-means was used for picture segmentation, the grey level cooccurrence matrix was used for feature extraction, and a multi-class SVM method with a linear Kernel was used for classification. They recorded a 95,99 % accuracy rate. The goal of the work by A. Singh and H. Kaur,(21) which was published in the IOP Conference Series: Materials Science and Engineering in 2021, was to use machine learning techniques to identify and categorize potato plant leaf diseases. Three algorithms' accuracy performance, namely Naive Bayes (NB), Support Vector Machine (SVM), and Convolutional Neural Network (CNN), were compared in the study. The CNN model, which had an accuracy of about 91 %, fared better than the other two models, according to the data. This work emphasizes the potential of CNN models in precisely identifying and diagnosing plant illnesses, particularly in potato crops, which can result in more efficient disease management techniques and improved agricultural yields. The authors,(22) trained and tested four ML classifiers, including decision tree (DT), k-nearest neighbor (KNN), support vector machine (SVM), and random forest (RF), on a dataset of 1 200 images of healthy and diseased potato leaves. The results of their study showed that the RF classifier outperformed the other classifiers, achieving an accuracy of 99,83 % in detecting and classifying potato leaf diseases. The SVM classifier came in second, achieving an accuracy of 98,50 %, while the DT and KNN classifiers achieved accuracies of 94,00 % and 91,50 %, respectively. The authors concluded that the RF classifier was the most effective for detecting and classifying potato leaf diseases, while SVM was also a strong performer. The study highlighted the potential of ML algorithms in improving the accuracy and efficiency of disease detection in agriculture, which could lead to more effective disease management strategies and ultimately, better crop yields. The findings of this study could be beneficial for farmers and agricultural workers in detecting and addressing plant diseases in a timely manner, thereby improving agricultural productivity and contributing to food security. Maintaining high-quality agricultural production depends on the timely diagnosis of plant diseases. Using photos of afflicted plants, machine learning, and deep learning approaches have recently demonstrated encouraging results in the precise identification and classification of plant diseases. T.-Y. Lee et al.,(23) employed convolutional neural networks (CNNs) to target the detection of potato leaf diseases in this study. The scientists made use of a dataset that included pictures of potato leaves that were taken in diverse fields with varying degrees of disease severity. They evaluated the effectiveness of numerous CNN models, including VGG16, ResNet, GoogleNet, and AlexNet. TensorFlow, a deep learning framework, was used to train and evaluate the models. The ResNet model fared better than the other models, with an accuracy of 97,5 %, according to the data. The scientists also assessed the effectiveness of the ResNet model on a real-time disease detection system for potato leaves, which was successful in accurately classifying potato leaf pictures into healthy and sick groups. The study shows that deep learning methods, notably CNNs, have the ability to accurately detect and categorize plant illnesses. By enabling farmers and agricultural workers to quickly identify and address plant illnesses, the discoveries might have a huge impact on the agriculture sector by improving crop quality and production. Using a collection of leaf image data, the research(24) developed a deep interpretable architecture for the categorization of plant diseases. To achieve high accuracy and interpretability, the design blends a convolutional neural network (CNN) with decision trees. The photos are processed by CNN to extract features, which are then put into decision trees to create classifications based on a predetermined set of criteria. The suggested design classified plant illnesses with a high accuracy rate of 99,8 % on the dataset, proving its efficacy. In order to eliminate any extraneous items, the authors,(25) preprocessed a collection of photos of healthy and ill plants from diverse sources. Then they put their CNN design into practice, which consists of two fully connected layers, three max-pooling layers, and five convolutional layers. The model was developed and evaluated using the dataset, and the results indicated a detection and classification accuracy of 94,5 % for plant illnesses. They discovered that the model concentrated on particular plant parts, such as the leaves and stems and that various illnesses were connected to various CNN activation patterns. Overall, the study demonstrates that CNNs might be a useful tool for identifying and categorizing plant diseases. The accuracy of the model may be further increased by using visualization methods like Grad-CAM, which can reveal information about how the CNN generates its predictions. The portions of the photos that CNN predicted as being most important were visualized by the authors using a method called Grad-CAM. They were better able to comprehend how CNN was able to distinguish between healthy and unhealthy plants as a result. In,(26) The authors of this research suggest a Convolutional Neural Network (CNN) model for identifying and categorizing plant diseases. The activation function of the CNN model has been optimized to increase classification accuracy. 3 680 photos of healthy and sick leaves from four distinct plant species make up the study's dataset. By making the photos grayscale, normalizing the pixel values, and shrinking them to a fixed size of 256 × 256, the authors pre-process the dataset. A training set, a validation set, and a testing set are created from the dataset.
Two convolutional layers with 32 and 64 filters each comprise the first two layers of the CNN model, followed by two fully connected layers with 256 and 128 neurons each. The Sine Linear Unit (SiLU) function was chosen as the study's optimal activation function. ReLU, Leaky ReLU, and Exponential Linear Unit (ELU) are three additional common activation functions that the authors evaluate the performance of their model. The outcomes demonstrate that, with an overall accuracy of 98,45 %, the CNN model with the SiLU activation function surpasses the other models. The authors also compare their model with other cutting-edge algorithms for identifying and categorizing plant diseases.
Regarding accuracy and F1 score, the suggested model performs better than the other models. In order to identify and categorize plant diseases, the authors suggest a CNN model with an optimized activation function. The outcomes show how the suggested methodology is successful in reaching high accuracy. Tiwari, Joshi, and Dutta et al.(27) introduced a dense convolutional neural network (DCNN) based plant disease detection and classification system. Using photographs of leaves, the technique is intended to categorize 12 different plant diseases. The authors used the Plant Village dataset, which contains 54 305 photos of leaves from 26 different crop species. The preprocessing of the dataset involved scaling the photographs and utilizing data augmentation methods to expand the training set. The authors then created DCNN architecture with four dense blocks and a transition layer. A 5-fold cross-validation strategy was used to train and test the system. The authors also assessed the system's performance in terms of its accuracy, recall, and F1 scores. The findings demonstrated the system's efficiency in disease identification and classification, which showed high values for all metrics. The performance of the suggested system was also evaluated against other cutting-edge models, such as VGG16, ResNet50, and Inception-v3. The outcomes revealed that the DCNN-based system performed better than all other models, illuminating its efficiency in identifying and categorizing plant diseases. To help farmers reduce crop losses, the scientists concluded that their technique may be effective for early identification of plant diseases. Mahum and Munir et al.(28), use an effective deep learning model to present a novel framework for detecting potato leaf disease in their study. The three key phases of the proposed framework are data gathering, pre-processing, and classification. A dataset of 5 000 photos of healthy and sick potato leaves was gathered for data collection. To improve the quality and lower noise, the photos underwent pre-processing. The authors employed data augmentation techniques to expand the dataset and lessen overfitting. The authors then advanced a powerful deep-learning model built on the Inception-v3 architecture. Transfer learning, which entails taking a pre-trained model and re-training it on a fresh dataset, was used to optimize the model. The authors trained the model using the gathered dataset and attained a 98,5 % accuracy rate. When they compared the accuracy of their model's performance to that of other cutting-edge models, they discovered that it performed better. The authors ran tests on an Android mobile application to further confirm the efficacy of their suggested architecture. The program was created with TensorFlow Lite and could quickly identify illnesses affecting potato leaves. The accuracy of the smartphone application, which the scientists tested using 100 photos of potato leaves, was 96 %. The authors conclude that their suggested framework is reliable and effective for identifying potato leaf disease. High classification accuracy was made possible by deploying an effective deep learning model built on the Inception-v3 architecture with transfer learning. The validation it received on a mobile application demonstrates the framework's ability to be used practically in the field. According to the authors, the suggested framework may include more crops and plant diseases for early identification and prevention. The detection and classification(43) of plant diseases using machine learning approaches is the subject of this paper's systematic review by Deepkiran Munjal et al.(29). Based on predetermined inclusion criteria, the author examined several scientific databases and chose 25 papers for evaluation. The research, which was published between 2016 and 2020, employed a variety of machine-learning techniques for plant disease identification and classification, including support vector machines (SVM), convolutional neural networks (CNN), and decision trees (DT). A comparison of articles based on the deep learning models is given in table 1.
|
Table 1. Comparative analysis of recent CNN based methods |
|||
|
Ref. |
Dataset |
Proposed Technique |
Objective |
|
(30) |
A dataset consisting of 500 images, categorized into 10 different classes, was gathered directly from field environments. |
The primary application of the article's approach is the detection of diseases in rice plants. By using deep transfer learning, the model can analyze images of rice plants and classify them into different disease categories. interventions and preventive measures. |
The article likely employs deep transfer learning techniques for rice plant disease detection. Transfer learning involves leveraging pre-trained models on large-scale datasets and fine-tuning them for a specific task, in this case, rice plant disease detection. |
|
(31) |
A total of 120 images utilized consisting of 3 categories. |
SIFT features-based classification |
The article's goal is to provide a way for automatically spotting and identifying illnesses in photos of rice plant leaves. The scientists hope that by using this method, they can help with the early detection and control of illnesses that damage paddy plants, improving the health and yield of the crop. |
|
(32) |
1500 photos of diseased and healthy rice and potato crop leaves were utilized in addition to 5932 photographs of rice. |
CNN model is proposed for the detection of potato leaves disease detection. |
The study's stated aim is to create a deep learning-based computer vision method, especially a Convolutional Neural Network (CNN) model, for the precise categorization of plant diseases in photographs of the leaves of rice and potato plants. The objective is to offer a low-cost, easily used, and trustworthy method for determining plant diseases without the need for laboratory testing or professional judgement. |
|
(33) |
1239 images with various resolutions captured with a smartphone camera at all phases (early, late) are input into a deep detector to identify illnesses in maize plants. |
Specifically using Faster R-CNN (Region-based Convolutional Neural Network) with the ResNet50 architecture, the study intends to integrate image recognition techniques and deep learning approaches in order to reliably diagnose the illnesses from real-time photographs. The suggested system measures the accuracy rate of the detection model to assess the model's efficacy. |
The goal of this study is to create an automated detection method for real-time picture analysis using deep learning and image processing techniques. The Northern Leaf Blight, Common Rust, and Cercospora leaf spots are the three common maize plant diseases that the algorithm tries to recognise and classify. The goal is to improve disease detection and enable prompt intervention to safeguard the maize crop, which serves as a staple food and a raw material for several businesses. Maize is an important crop in India. |
|
(34) |
Total of 115 images are utilized consisting of 33 brown spot images, 42 leaf smut images, and 40 bacterial leaf blight images. |
A method is proposed based on the DenseNet169-MLP model. To conduct rice plant disease classification, the final layer is replaced by the MLP technique, and the feature extractor is the pretrained DenseNet169 method. |
The goal of the project is to combine MLP (Multilayer Perceptron), a kind of artificial neural network, with DenseNet169, a deep learning architecture, to develop an efficient classification model for illnesses of rice plants. The precise classification of rice plant diseases is handled by the DenseNet169-MLP model, which was created with this objective in mind. |
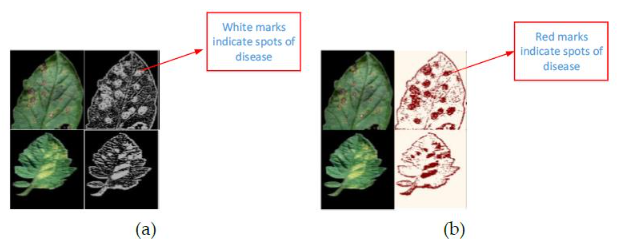
Figure 1. Segmentation using a binary threshold algorithm(14)
The analysis discovered that CNN-based models, which have high accuracy rates in most experiments, are the most often utilized deep learning technique for classifying and detecting plant diseases. In several cases, models built using SVM and DT also did well. Transfer learning and the usage of data augmentation approaches have been demonstrated. Machine learning models have been demonstrated to perform better when data augmentation methods and transfer learning are used. The accuracy of the models is also influenced by the selection of elements taken from the plant photos, such as texture, color, and form. The lack of standardization in image capture and labelling, the restricted availability of large-scale datasets, and the difficulty of diagnosing diseases in their early stages are only a few of the limits and difficulties the author noticed with the current research.
Proposed methodology
In this part, we'll outline the procedures used in our experimental setup to classify plant illnesses using various machine learning techniques and a unique deep learning model. The three main stages of our suggested methodology are data collection, image pre-processing, feature extraction, and classification. The next parts provide a brief overview of the experimental configuration, the dataset, and the key stages of our suggested methodology:
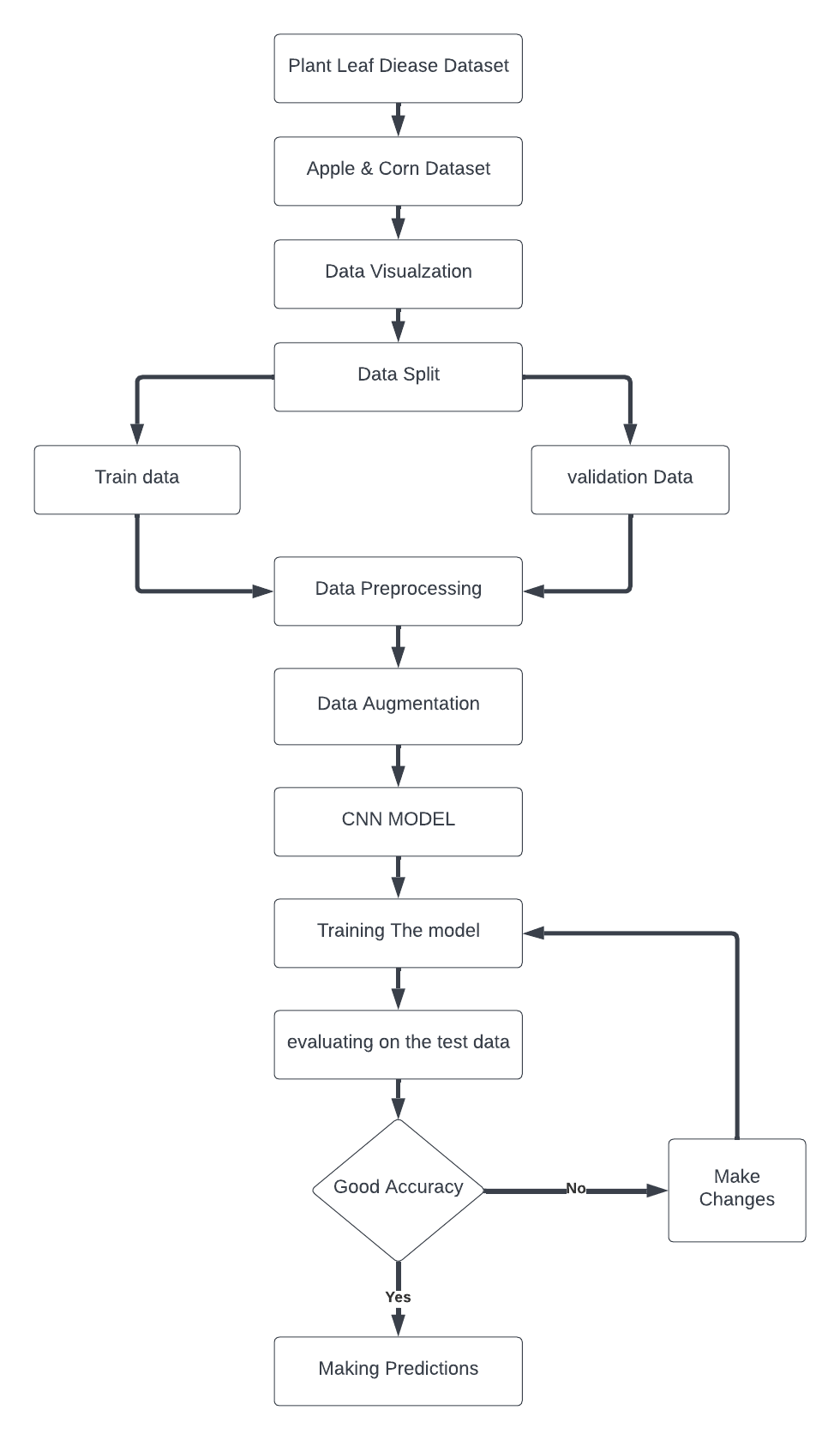
Figure 2. Proposed Method Block Diagram
System Background
Diagnosing plant diseases is difficult because of the many species' apparent similarities and profound structural differences. Early detection and categorization are essential to halt the spread of disease in crops. Deep Convolutional Neural Networks (CNNs) have shown promise in identifying diseases from images. This study investigates several machine learning and deep neural network methods for identifying plant diseases from photos of afflicted plants. The main goal is to create a CNN model and highlight how crucial hyperparameters are for obtaining correct results.
Experimental Setup
In this section, a brief description of the experimental setup will be discussed. Experiments are performed on the system having 8GB RAM, Core i5, 7th generation. Simulations of the research are performed on the Python language version 3,8 in Google Colab with a GPU processor.
Dataset
The PlantVillage dataset(35) is utilized in this work, which contains 61 486 images with 39 different categories of healthy and diseased leaves related to 14 plant species. The image size is adjusted to 224× 224×3. The dataset is publicly available for research purposes. We used 2 plants, Apple and Corn, for our experiment, containing 7023 images of 8 different classes. Some sample images of the PlantVillage dataset are shown in figure 3.
Image pre-processing
This section goes into great depth on the proposed method's initial step. The developed CNN model requires a large amount of data for training to reduce the possibility of over-fitting and increase the variety of the data set. For this reason, a variety of data augmentation techniques, such as horizontal flip, vertical flip, upside down, downside up, and additional noise, were applied to the training set. The Python Image Data Generator function of the Keras package was used to enhance the data. To decrease computing costs, scale transformation was first employed. Pixel values were ranged from 0 to 1 using the parameter value (1/255). We used a 25° rotation transformation. The width and height shift range parameters were set to 0,1. Additionally, a shear angle of 0,2 was used. The image was enlarged using a random zoom transformation with a 0,2 zoom range value. Flipping horizontally was done. We used a brightness range of 0,5 to 1,0. The fill mode was closest, and the channel shift transformation utilized a 0,05 value.
Feature Extraction
In the field of object recognition, detection, and classification, researchers use different methods to extract features. In our study, we specifically focus on employing Convolutional Neural Network (CNN) models for feature extraction. This process involves two main stages: feature extraction and feature optimization.
During feature extraction, we use various CNN models to capture and interpret complex patterns within image data. These models are designed to automatically extract meaningful features from input images, enabling the network to comprehend the unique characteristics of different objects or elements within the images.
Following feature extraction, feature optimization involves fine-tuning the network's hyperparameters. This requires careful manual adjustment based on different ranges for each hyperparameter. Parameters such as learning rates, batch sizes, and regularization techniques are meticulously tuned within specified ranges to optimize the network's performance. This ensures that the network effectively learns and represents the most relevant features for accurate object recognition and classification.
Our goal in exploring these phases is to enhance the network's capability to extract and optimize features, ultimately contributing to more precise and reliable object recognition and classification within the scope of our research.
The features extraction and features optimization phases using manual hyper-parameter tuning based on different ranges of each hyper-parameter are explained in the following sections:

Figure 3. Sample Images of the Dataset
Classification Algorithms
K-Nearest Neighbors (KNN), decision trees, and support vector machines (SVMs) are three well-liked classification techniques used in machine learning. The significance and mathematical underpinnings of algorithm are explained as follows:
a) Support Vector Machines (SVM)
Data that can be divided into linear and non-line categories may be handled using SVMs, which are efficient classifiers. SVM aims to identify the hyperplane that best separates the different classes in the dataset. SVM does this by extending the distance between each class's support vectors, or closest points, from the hyperplane. SVM can also handle high-dimensional data by translating high-dimensional data into a higher-dimensional space where the classes are separable using kernel approaches. Mathematically, the optimization problem of SVM can be written as:
min_(w,b,ξ ) 1/2 IIwII^2+C∑_(i-1)^n ξi (1)
Subject to the:
y_i (w^T x_i+b)≥1-ξ_i, i∈(1,2,…,n)
ξ_i≥0,∀i∈(1,2,…,n)
Where, and are the coefficients of the hyperplane, is the slack variable representing the degree of misclassification of the sample, is the label of the sample, and is a regularization parameter that controls the trade-off between maximizing the margin and minimizing the misclassification.
b) Decision Tree (DT):
A Decision Tree is a simple representation for classifying examples. It is a Supervised Machine Learning where the data is continuously split according to a certain parameter. It consists of set of nodes which test for the value of a certain attribute, edges or branch which correspond to the outcome of a test and connect to the next node or leaf, and leaf nodes that represent class labels and predict the outcome. Entropy is the key idea in this algorithm which helps to identify the function or attribute which provides the most information on a class. The objective is to decrease the entropy level from the root of the tree to the root node of leaf. The equation is given below:
E(s) =∑(i=1)^k P(i) Log2 (P(i)) (2)
c) Random Forest (RF):
Random Forest is indeed a flexible learning machine technique which is suitable for a range of applications including classification and regression problems. It is an ensemble technique, which means that a random model of forest consists of a large number of tiny decision-making trees called estimators each producing their own predictions. The random forest algorithm integrates the estimators’ predictions to create a more precise forecast. The Gini index is used to select the node on a decisions tree branch.
Gini =1-∑(i=1)^k Pi^2 (3)
Naive Bayes is a classification technique that is
one of the simple yet effective algorithm, it classify observations using Bayes
Theorem. Its concept is based on an assumption of independence between
predictors. In simple terms, a Naive Bayes classifier assumes that the presence
of a particular feature in a
class is unrelated to the presence of any other feature. The main formula of
Naive Bayes is given below:
P(Y⁄X)=(P(X⁄Y)P(Y))/(P(X)) (4)
d) K-Nearest Neighbor (KNN):
The k-nearest neighbor approach varies from the
other methods discussed in that it uses the data directly for classification
rather than first developing a
model. As a result, no model design details are needed, and the only variable
in the model is k parameter, which specifies the number of nearest neighbors to
include in the estimation of class membership. It based on a similarity measure
(e.g., distance functions). The model can be made more or less flexible by
changing k. The advantage of k-nearest neighbors over other algorithms is that
the neighbors can explain for the classification result.
d( Pi,Qj)=√(∑_(r=1)^n(Pir-Qjr)) (5)
e) Logistic Regression (LR):
A logistic regression model specifies that an adequate function of the fitted probability of the occurrence of the event is a linear function of the observed values of the relevant explanatory variables. It is a probability model that is expressed in terms of the likelihood of an event (Y) occurring under a given set of conditions (X):
P=e^(β0+β1 x)/(1+e^(β0+β1 x)) (6)
The main benefit of this model is that it can generate a simple probabilistic classification formula. In addition, it is useful for predicting the existence or absence of a characteristic or outcome based on the values of a set of predictor variables.
f) XGBoost:
XGBoost stands for Extreme Gradient Boosting is an ensemble learning technique to build a strong classifier from several weak classifiers in series.
Boosting algorithms play a crucial role in dealing with bias-variance trade-offs.
Unlike bagging algorithms, which only control for high variance in a model, boosting controls both the aspects (bias and variance) and is considered to be more effective. it is a specific implementation of the Gradient Boosting method that uses more accurate approximations to find the best tree model. It uses anumber of tricks that make it exceptionally successful, especially with structured data, the most important are the partial derivatives of the loss function which provides more information about the direction of the gradients and how to reach the minimum of our loss function. XGBoost uses the second orderderivative as an approximation.In addition; it uses advanced regularization (L1& L2), which improves the generalization of the model. XGBoost has additional advantages which is the training is very fast and can be parallelized and distributed between clusters. Besides the use of traditional machine learning models in this study, a deep learning approach is developed based on a custom convolutional neural network model since they can achieve to better results and have good accuracy than a traditional machine learning models.
g) Convolutional neural networks (CNNs):
Convolutional neural networks (CNNs) represent a distinct form of neural networks that utilize convolution, differing from traditional matrix duplication in at least one layer. In simpler terms, CNNs are specialized artificial neural networks tailored for image recognition and processing, specifically optimized for pixel data.
A typical convolutional neural network comprises an input layer, hidden layers, and an output layer. Within a feed-forward neural network, intermediate layers are termed "hidden" since their inputs and outputs are concealed by the activation function and final convolution. In the case of a convolutional neural network, these hidden layers involve performing convolutions, often involving a dot product between the convolution kernel and the input matrix. This product generally takes the form of the Frobenius inner product, with the ReLU function commonly serving as its activation function. As the convolution kernel moves across the input matrix for the layer, the convolution operation produces a feature map, subsequently contributing to the input of the subsequent layer. This process is succeeded by additional layers such as pooling layers, fully connected layers, and normalization layers.
The quantitative results of the proposed method based on the optimized CNN features are given in the following section.
RESULTS AND DISCUSSION
The findings of the studies are provided and thoroughly explored in the results section. The main goal of the study is to assess how altering convolutional neural networks (CNN) hyperparameters affects prediction accuracy. A popular deep learning architecture for image identification applications is CNN. A model trained for one job can be utilized as a starting point for training on another task using the machine learning approach known as transfer learning. The CNN models are trained using various hyper-parameter values adjusted manually, and the models' accuracy is assessed.
Experiments based on CNN features
In this research section, Experiments based on the deep learning features are discussed. The CNN models are trained using different hyperparameter values with manual configuration, and the models' accuracy is assessed. The research's conclusions will aid in choosing the CNN model's ideal hyperparameter values, increasing the accuracy of image recognition tasks. Additionally, comparing pre-trained CNN models that have undergone transfer learning will provide light on the efficiency of transfer learning in enhancing CNN model performance. A brief description of the experiments performed using the CNN model is given in the following sub-sections:
Test 1: Results based on the proposed CNN Model 0
The supplied deep learning model, model_0, features a max-pooling layer with a pool size of (2, 2) after each set of four convolutional layers. Filter sizes for the last three convolutional layers are 32, 64, and 128 respectively. The convolutional layers' filter size starts at 16 and increases in multiples of 8. The result is then transmitted to a layer called "flatten," which creates a one-dimensional array using the information from the layers that came before it. The output of the flattened layer is then coupled to three sets of fully connected layers, each set consisting of a fully connected layer and a dropout layer. The dropout layer's value of 0,3 randomly sets input units to 0 with a frequency of rate at each step during training in order to prevent overfitting. To keep the total constant across all inputs, non-zero inputs are scaled up by 1/ (1 - rate). The output of the last fully connected layer is sent into the output layer, which uses the softmax function as the activation function to anticipate a multinomial probability distribution. Figure 4 depicts the layered architecture of this model.
The limited availability of images necessitated various techniques such as rescaling, rotation, shifting, zooming, and flipping to expand the training dataset. The Adam optimizer was used to train the model across 50 epochs with a batch size of 32 and a 128 by 128 picture shape. After training, the model predicted test accuracy to be 97 % and test loss to be 8 %. The batch size, picture shape, optimizer, and epochs are only a few of the variables that significantly impact the model's performance. While the image size identifies the dimensions of the input photo, the batch size establishes the number of samples every training cycle. While the number of epochs controls how often the model iterates over the whole training dataset, the optimizer specifies the optimization approach used to change the model's parameters during training.
The proposed model 0 utilizes convolutional and fully connected layers with dropout regularization and softmax activation in the output layer to achieve good accuracy in photo categorization challenges. The model's performance received a significant boost, and overfitting was avoided, by expanding the training dataset. The model's hyper parameters had a significant impact on its performance. After 50 training rounds using an Adam optimizer with a 32-batch size and a 128 × 128 picture shape, the model had a test accuracy of approximately 97 %.
Test 2: Results based on the proposed CNN Model 1
By adjusting the hyperparameters and adding new layers to the architecture of the prior model in this experiment, we were able to significantly boost performance. First, in order to enable the model to extract additional characteristics from the picture, we extended the image shape from 128 × 128 to 224 x 224. In the third experiment, we added a dropout layer with a 0,3 drop rate, a Conv2D layer with 256 filters, a MaxPooling layer with a pool size of (2, 2), and a Dense layer with 256 neurons to improve the model's capacity to extract features. We trained the model for 50 epochs using data augmentation methods comparable to those employed for the prior model. Following the completion of training, we evaluated the model's accuracy and loss on the test dataset, which was found to be 97,6 % and 7 %, respectively. This is a modest improvement over the prior model.
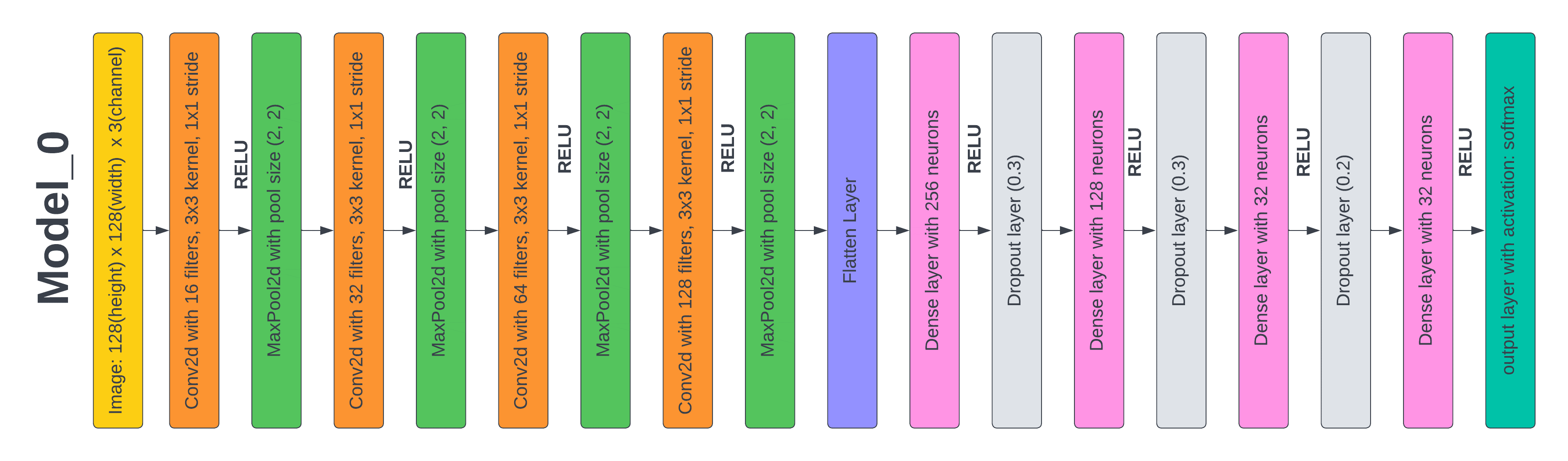
Figure 4. Layered Architecture of Model 0
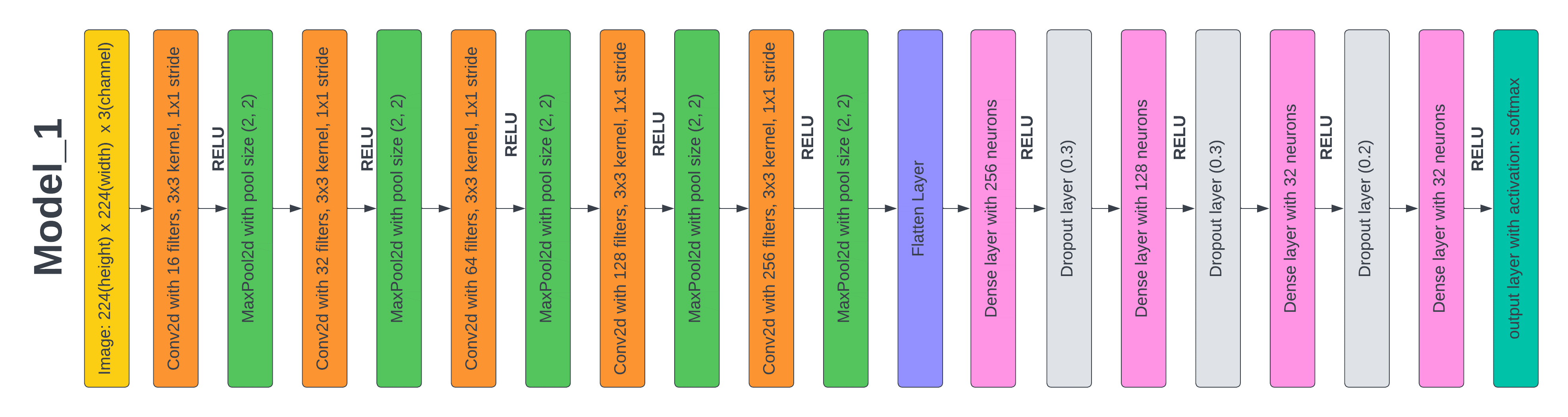
Figure 5. Layered Architecture of Model 1
By enabling more thorough feature extraction capabilities and limiting overfitting by including dropout layers, these adjustments to the hyperparameters and including layers have
enhanced the model's performance. This model's increased accuracy suggests it is more adept at identifying and categorizing various image types. However, it is vital to remember that these findings might only generalize to some datasets and that more testing is necessary.
Test 3: Results based on the proposed CNN Model2
In the third experiment, we adjusted the hyperparameters of model_1 to examine their impact on the model's performance. The initial modification involved increasing the batch size from 32 to 64. The batch size refers to the data processed before each model update during training. While a larger batch size may lead to more accurate weight updates, it can consume more memory and slow down the training process. To determine if the new batch size impacted the model's accuracy, we trained the model for 50 epochs. We raised the first filter's size from 16 to 32, and in the succeeding convolutional layers, we steadily increased the filter size by a multiple of 8. The network may learn more complicated characteristics from the input photos by adding additional filters to each layer. We discovered that the accuracy on the test dataset declined marginally to 97,3 %, and the loss stayed at 7 % after training the model for 50 epochs with these improvements. This indicates that raising the batch size did not enhance the model's performance and that increasing the filter size could have caused overfitting, which resulted in a tiny drop in accuracy. Consequently, we concluded that the model_1 original architecture with its hyperparameters was the most effective for the dataset.
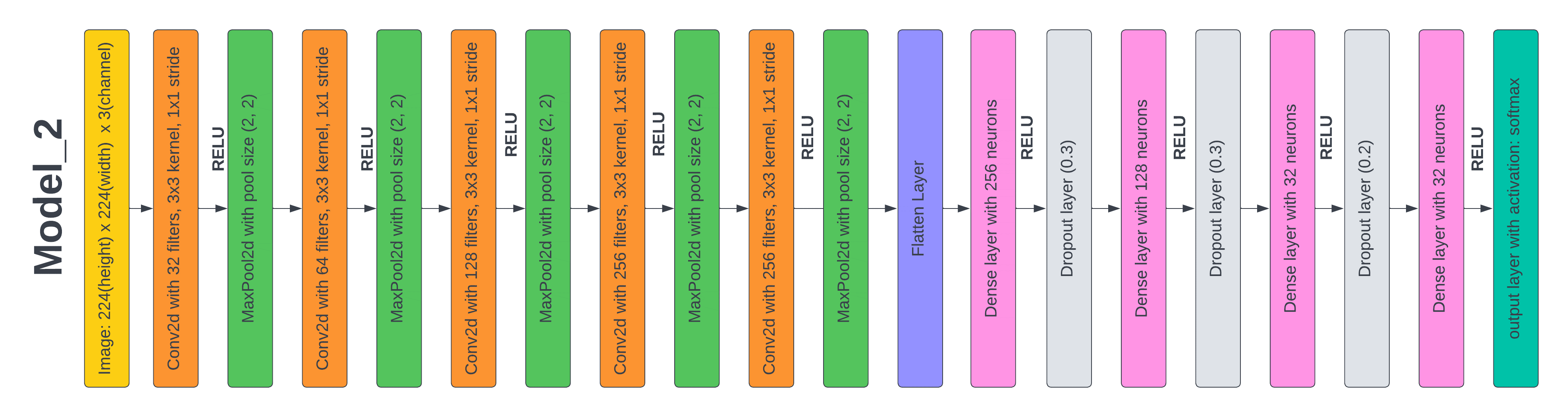
Figure 6. Layered Architecture of Model 2
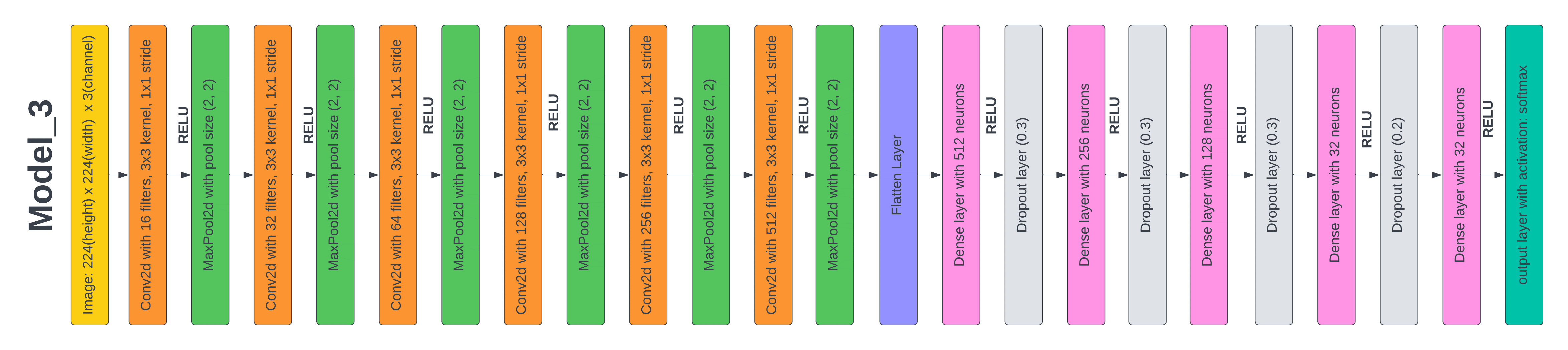
Figure 7. Layered Architecture of Model 3
Test 4: Results based on the proposed CNN Model 3
We used the model_2 as the basic model for the model_3 and adhered to the same architecture. However, the filter sizes used in the conv2D layers have seen some adjustments. For each conv2D layer, we modified the filter size specifically, starting at 16 and increasing it by a multiple of 8, ensuring that the filter size used in the current layer is equal to or greater than the one used before. This was done to examine the results of beginning with a smaller number and reducing the filter size. The model has a 96 % accuracy and a 19 % loss on the test data after 50 training epochs. This one has the lowest accuracy and the biggest loss out of all the models we've tried. This result indicates that starting with a lesser value and reducing the filter size has a detrimental impact on the model's performance. It is important to note that the individual issue and dataset being utilized significantly influence how hyper parameters affect a model's performance. As illustrated in figure 7, the model's architecture represents the culmination of our current achievements.
Test 5: Results based on the proposed CNN Model 4
In our earlier research, we noticed that accuracy decreased as the batch size in the model rose. We chose to carry out another experiment in which we merely altered the batch size of the model to further analyze this. We reduced the batch size in this model_4 from 64 to 32. We just altered the batch size parameter, leaving the remainder of the model design the same as model_2. The model_4 had an accuracy of 98,2 % and a loss of 6 % on the test dataset after 50 epochs of training.
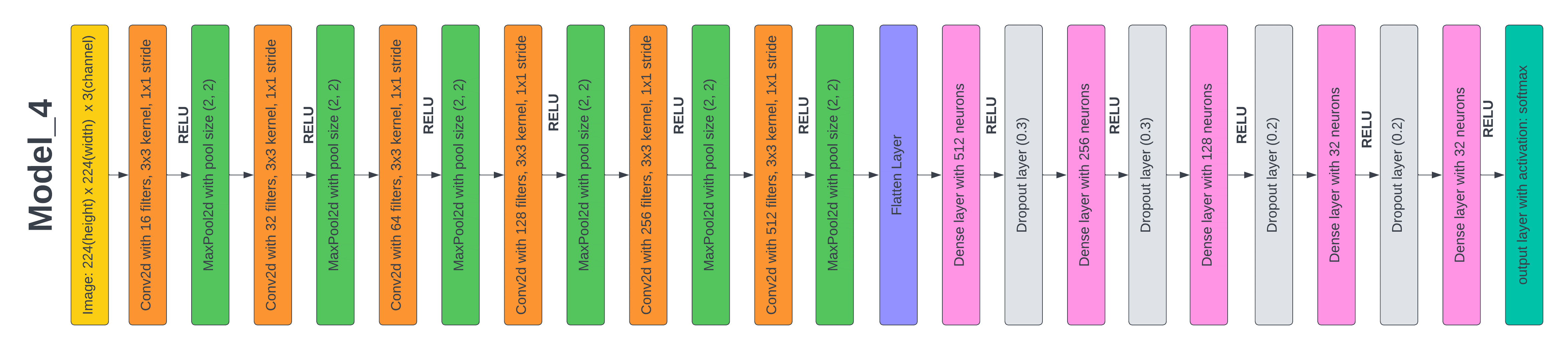
Figure 8. Layered Architecture of Model 4
|
Table 2. Accuracy of classical machine learning algorithms |
|
|
Machine Learning Models |
Accuracy |
|
KNN |
82,20 % |
|
SVM |
87,75 % |
|
Logistic Regression |
86,83 % |
|
XGBoost |
84,83 % |
|
Naïve Bayes |
63,84 % |
|
Decision Tree |
70,39 % |
|
Random Forest |
72,59 % |
|
Table 3. Quantitative results comparison of modified CNN_models |
|||||
|
Performance Measures |
Model_0 |
Model_1 |
Model_2 |
Model_3 |
Model_4 |
|
Accuracy |
96,89 % |
97,60 % |
97,31 % |
95,30 % |
98,02 % |
|
Precision |
96,20 % |
97,30 % |
97,00 % |
96,20 % |
97,30 % |
|
Recall |
96,00 % |
96,80 % |
96,30 % |
94,60 % |
97,50 % |
|
F1 score |
95,50 % |
97,20 % |
96,70 % |
95,20 % |
97,30 % |
Quantitative Results Comparison of all Modified Models based on CNN
We assessed the performance of five distinct models for image classification tasks, ranging from the basic model_0 to the complex model_4. The models were trained using a collection of images and the labels assigned to them to maximize accuracy while minimizing loss. The simplest model, Model_0, has one Conv2D layer and one Dense layer. The model was trained for 50 epochs using a batch size 32 and pictures of 128 by 128 shape. Model_1 improved over Model_0, with additional layers and hyperparameters set for better performance. The model had a Conv2D layer with 256 filters, a max pooling layer, a Dense layer with 256 neurons, and a dropout layer with a rate of 0,3. The image size was increased to 224 × 224.
With the same batch size and 50 epochs, Model_2 was modified by increasing the filter size and batch size. Model_2 had an accuracy of 97,6 % and a loss of 7 % on the test dataset, outperforming Model_0. The batch size was also raised to 64. The model could have performed better, with an accuracy of 97,3 % and a loss of 7 % on the test dataset. Model_3 changed the filter sizes over Model_2, utilized as a basis model. We reduced the filter size, which started at 32 in Model_2 and was steadily reduced by a multiple of 8 in succeeding Conv2D layers. The model fared the poorest, attaining an accuracy of 96 % and a loss of 19 % on the test dataset after being trained for 50 epochs with a batch size of 32. Model_4 just changed the batch size while using Model_2 as a basis model. In Model_4, the batch size was reduced from 64 to 32. The best-performing model, trained for 50 epochs, exhibited an accuracy of 98,2 % and a loss of 6 % on the test dataset, outperforming all other models.
In conclusion, the experiments have demonstrated that important variables such as image shape, batch size, and filter size substantially impact the performance of machine learning algorithms for image classification tasks. The studies also showed that while raising the batch size might result in a loss of accuracy, decreasing the filter size can have a detrimental impact on the model's performance. The studies demonstrate that the model's accuracy may be improved by increasing the number of layers and neurons in the Dense layers. Table III presents the findings, while Figures 9, 10, and 11 display each trial's confusion matrix and ROC curves. Model_4 was significantly better than the other models in terms of accuracy, according to the results.

Figure 9. Confusion Matrices of all the Models
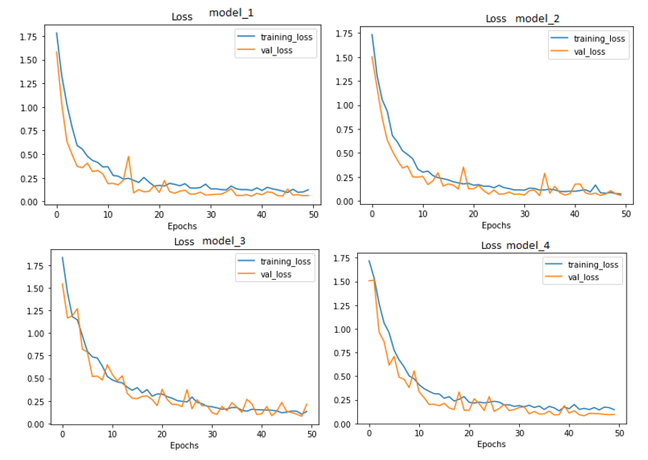
Figure 10. Loss ROC Curves of all Modified Models
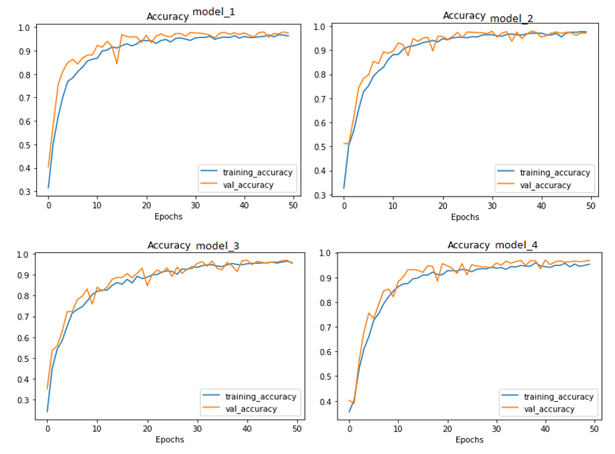
Figure 11. Accuracy Curve of all Modified Models
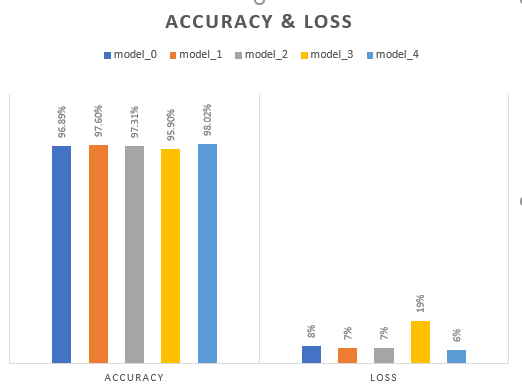
Figure 12. Accuracy and Loss graphs of all modified models based on CNN
Comparison whit existing method
In this section, we conduct a comprehensive comparison between our proposed methodology and traditional machine learning algorithms, all applied to our dataset. Furthermore, we benchmark our approach against some state-of-the-art techniques in the field.
To commence, we scrutinize supervised machine learning algorithms, including Random Forest (RF), Logistic Regression (LR), XGboost, Decision Tree (DT), Naïve Bayes (NB), and Support Vector Machine (SVM). These represent a subset of well-established supervised algorithms for machine learning. We couple these algorithms with advanced image processing techniques and meticulously analyze the resultant algorithmic outputs, striving to identify the most adept solution for plant disease classification. This performance surpasses all other candidates in our comprehensive evaluation, as demonstrated in table 2.
In other hand, Several performance metrics were used to measure the robustness of our proposed models .The comparison of results in table 3 shows that the model 4, having same number of epochs in other models, out performs all other models as its average precision, recall, specificity and F1_measure (F1-Score) are 98,2 %, 97,3 %, 97,5 % and 97,3 % respectively. We can conclude from this experiment that the architecture and values of hyperparameters used in model 4 gives the best results on apple and corn dataset since it is able to classify images of 8 different classes of apple and corn. In addition,
the accuracy decreased by increasing the number of batch
size. Furthermore, the proposed method’s performance was compared to the corn and apple leaf disease detection existing techniques from the literature to demonstrate the generalization of the proposed approach. The results showed that the proposed method achieved the highest accuracy compared to existing studies. The proposed CNN model (model 4) outperformed the existing studies as (36) achieved 92,85 % accuracy.
The authors of (37) reported 92,43 % accuracy with superpixel-based disease classification method using end-to-end CNN (convolutional neural network) architecture with higher computational cost. While (38) achieved an average accuracy of 97,2 % by using transfer learning for automatic apple disease identification and classification on them prepared dataset. In addition,(39) achieved an accuracy of 98 % in classifying the Corn leaf images into four different categories. Also,(40) achieved an accuracy of 98 % by concatenating CNN-extracted features with ANN and the proposed method in(41) identify the crop species with 96,76 % accuracy. Thus, the proposed CNN model dominated the existing techniques, thus achieving 98,02 % accuracy.
In table 4, we have performed further comparisons between our proposed method model _4 and other existing techniques. Our findings not only demonstrate that model_4 consistently outperforms a significant portion of prior research in predicting and classifying plant diseases but also highlight its superiority in terms of accuracy and its ability to achieve these results with a reduced number of parameters.
|
Table 4. Comparison of our proposed method with existing techniques |
||||||
|
Ref |
Year |
Total Images |
Methodology |
Plant Diseases/ Deficiency |
Classes |
Accuracy |
|
(31) |
2016 |
120 |
SIFT features, segmentation approach, SVM for classification |
Potato |
3 |
91,1 % |
|
(30) |
2017 |
500 |
Deep Learning approach (CNN) |
Potato |
10 |
95,38 % |
|
(42) |
2020 |
54000 |
Deep Learning (CNN) with optimized activation function |
Variety of plants |
36 |
97,82 % |
|
(34) |
2022 |
115 |
Deep Learning |
Potato |
3 |
97,68 % |
|
Proposed |
7023 |
Deep Learning Based on CNN |
Apple and Corn |
8 |
98,02 % |
|
CONCLUSION
The study explores deep learning and machine learning techniques for identifying plant diseases from images of affected plants. It encompasses data collection, image pre-processing, feature extraction, and classification, emphasizing the crucial role of hyperparameters in producing accurate results. With a dataset of 7023 images across 8 categories, the research utilizes CNN models and traditional machine learning algorithms to classify apple and maize leaf diseases. The CNN model outperforms other classifiers, achieving an accuracy of nearly 98 %. This surpasses previous machine learning-based methods and underscores the potential of deep neural networks for disease detection and image categorization. The study underscores the importance of carefully adjusting these parameters and highlights the significance of hyperparameters in obtaining precise findings. The proposed deep interpretable architecture for plant disease categorization represents a significant advancement in agricultural technology, potentially revolutionizing disease identification and management. Improving disease diagnostics could contribute to halting the spread of crop diseases and ensuring global food security.
In our future work, we plan to employ advanced hyperparameter tuning techniques such as genetic algorithms, grid search, random search, and Bayesian optimization algorithms to optimize the main and critical hyperparameters in our CNN model. This will enable us to efficiently fine-tune our model and identify the best architecture. By leveraging these techniques, we aim to streamline the process of identifying the most effective configuration for our CNN, ultimately enhancing its performance in diagnosing plant diseases.
Additionally, we intend to delve into segmentation techniques using pretrained models for a more in-depth analysis of plant diseases. By integrating segmentation methods into our analysis, we seek to achieve a more granular understanding of the affected areas in plant images. This approach will enable us to pinpoint and isolate specific regions within the plant images that are indicative of disease, thereby enhancing the overall accuracy and precision of our diagnostic processes. We believe that this deeper analysis will significantly contribute to the effectiveness of our disease identification system.
In addition to the optimization techniques and segmentation methods, our future work also involves the development of a mobile application. This application will be designed to provide detailed information about the affected areas within plant images, along with the nature of the diseases detected and recommended treatments. By incorporating a user-friendly mobile interface, we aim to empower users, including farmers and agricultural specialists, with a tool that can swiftly and accurately identify plant diseases in the field. This mobile application will serve as a practical and accessible resource, offering valuable insights and actionable recommendations based on our advanced disease analysis.
REFERENCES
1. T. Noulamo, A. Djimeli-Tsajio, J. P. Lienou, and B. Fotsing Talla, “A Multi-Agent Platform for the Remote Monitoring and Diagnostic in Precision Agriculture.” “IJCS_49_1_23”.
2. Kamilaris, F. X. J. C. Prenafeta-Boldú, and e. i. agriculture, "Deep learning in agriculture: A survey," vol. 147, pp. 70-90, 2018
3. Naim and N. I. Ghali, “New Optimization Algorithm Based on Venus Flytrap Plant,” IAENG Int J Comput Sci, vol. 48, no. 3, 2021.
4. T. Noulamo, A. Djimeli-Tsajio, J. P. Lienou, and B. Fotsing Talla, “A Multi-Agent Platform for the Remote Monitoring and Diagnostic in Precision Agriculture.”
5. Fuentes, S. Yoon, S. C. Kim, and D. S. J. S. Park, "A robust deep-learning-based detector for real-time tomato plant diseases and pests recognition," vol. 17, no. 9, p. 2022, 2017.
6. Ma and Y. Chen, “Attentive Enhanced Convolutional Neural Network for Point Cloud Analysis.”
7. J. Liu and W. Wu, “Automatic Image Annotation Using Improved Wasserstein Generative Adversarial Networks.,” IAENG Int J Comput Sci, vol. 48, no. 3, 2021.
8. Z. Iqbal et al., "An automated detection and classification of citrus plant diseases using image processing techniques: A review," vol. 153, pp. 12-32, 2018.
9. N. E. D. G. K. Water and P. G. T. P. O. J. J. A. S. EC, "Crop losses to pests," vol. 144, pp. 31-43, 2006.
10. K. Golhani, S. K. Balasundram, G. Vadamalai, and B. J. I. P. i. A. Pradhan, "A review of neural networks in plant disease detection using hyperspectral data," vol. 5, no. 3, pp. 354-371, 2018.
11. J. T. Lalis, “A new multiclass classification method for objects with geometric attributes using simple linear regression,” IAENG Int J Comput Sci, vol. 43, no. 2, pp. 198–203, 2016.
12. S. S. Chouhan, A. Kaul, U. P. Singh, and S. J. I. A. Jain, "Bacterial foraging optimization based radial basis function neural network (BRBFNN) for identification and classification of plant leaf diseases: An automatic approach towards plant pathology," vol. 6, pp. 8852-8863, 2018.
13. R. Long, D. Yang, and Y. Liu, “DiseaseNet: A Novel Disease Diagnosis Deep Framework via Fusing Medical Record Summarization.”
14. K. Eldahshan, H. Mancy, K. Eldahshan, E. K. Elsayed, and H. Mancy, “Enhancement Semantic Prediction Big Data Method for COVID-19: Onto-NoSQL.” [Online]. Available: https://www.researchgate.net/publication/354905909
15. Hernández-Flórez N. Breaking stereotypes: “a philosophical reflection on women criminals from a gender perspective". AG Salud 2023;1:17-17.
16. C. Koo et al., "Development of a real-time microchip PCR system for portable plant disease diagnosis," vol. 8, no. 12, p. e82704, 2013.
17. N. Tian and W. Zhao, “EAST: Extensible Attentional Self-Learning Transformer for Medical Image Segmentation.,” IAENG Int J Comput Sci, vol. 50, no. 3, 2023.
18. K. Salhi, E. M. Jaara, M. T. Alaoui, and Y. T. Alaoui, “Color-texture image clustering based on neuro-morphological approach,” IAENG Int J Comput Sci, vol. 46, no. 1, pp. 134–140, 2019.
19. N. E. M. Khalifa, M. H. N. Taha, L. M. Abou El-Maged, A. E. J. M. L. Hassanien, A. Big Data Analytics Paradigms: Analysis, and Challenges, "Artificial intelligence in potato leaf disease classification: a deep learning approach," pp. 63-79, 2021.
20. M. Islam, A. Dinh, K. Wahid, and P. Bhowmik, "Detection of potato diseases using image segmentation and multiclass support vector machine," in 2017 IEEE 30th canadian conference on electrical and computer engineering (CCECE), 2017, pp. 1-4: IEEE.
21. Singh and H. Kaur, "Potato plant leaves disease detection and classification using machine learning methodologies," in IOP Conference Series: Materials Science and Engineering, 2021, vol. 1022, no. 1, p. 012121: IOP Publishing.
22. Caero L, Libertelli J. Relationship between Vigorexia, steroid use, and recreational bodybuilding practice and the effects of the closure of training centers due to the Covid-19 pandemic in young people in Argentina. AG Salud 2023;1:18-18..
23. T.-Y. Lee, I.-A. Lin, J.-Y. Yu, J.-m. Yang, and Y.-C. J. S. C. S. Chang, "High efficiency disease detection for potato leaf with convolutional neural network," vol. 2, no. 4, p. 297, 2021.
24. M. Brahimi, S. Mahmoudi, K. Boukhalfa, and A. Moussaoui, "Deep interpretable architecture for plant diseases classification," in 2019 Signal Processing: Algorithms, Architectures, Arrangements, and Applications (SPA), 2019, pp. 111-116: IEEE.
25. M. Francis and C. Deisy, "Disease detection and classification in agricultural plants using convolutional neural networks—a visual understanding," in 2019 6th international conference on signal processing and integrated networks (SPIN), 2019, pp. 1063-1068: IEEE.
26. S. Y. Yadhav, T. Senthilkumar, S. Jayanthy, and J. J. A. Kovilpillai, "Plant disease detection and classification using cnn model with optimized activation function," in 2020 international conference on electronics and sustainable communication systems (ICESC), 2020, pp. 564-569: IEEE.
27. V. Tiwari, R. C. Joshi, and M. K. J. E. I. Dutta, "Dense convolutional neural networks based multiclass plant disease detection and classification using leaf images," vol. 63, p. 101289, 2021.
28. R. Mahum et al., "A novel framework for potato leaf disease detection using an efficient deep learning model," vol. 29, no. 2, pp. 303-326, 2023.
29. D. Munjal, L. Singh, M. Pandey, and S. J. I. J. o. S. I. Lakra, "A Systematic Review on the Detection and Classification of Plant Diseases Using Machine Learning," vol. 11, no. 1, pp. 1-25, 2023.
30. Y. Lu, S. Yi, N. Zeng, Y. Liu, and Y. J. N. Zhang, "Identification of rice diseases using deep convolutional neural networks," vol. 267, pp. 378-384, 2017.
31. K. J. Mohan, M. Balasubramanian, and S. J. I. J. o. C. A. Palanivel, "Detection and recognition of diseases from paddy plant leaf images," vol. 144, no. 12, 2016.
32. R. Sharma et al., "Plant disease diagnosis and image classification using deep learning," vol. 71, no. 2, pp. 2125-2140, 2022.
33. M. S. Kumar, D. Ganesh, A. V. Turukmane, U. Batta, and K. K. J. J. o. P. N. R. Sayyadliyakat, "Deep Convolution Neural Network Based solution for Detecting Plant Diseases," pp. 464-471, 2022.
34. Quiroz FJR, Oncoy AWE. Resiliencia y satisfacción con la vida en universitarios migrantes residentes en Lima. AG Salud 2023;1:09-09..
35. Shoaib, M., Hussain, T., Shah, B., Ullah, I., Shah, S.M., Ali, F., Park, S.H.: Deep learning-based segmentation and classification of leaf images for detection of tomato plant disease. Frontiers in Plant Science 13, 1031748 (2022)
36. T. O. Emmanuel. (2018). PlantVillage Dataset. Available: https://www.kaggle.com/datasets/emmarex/plantdisease
37. Sibiya, M., & Sumbwanyambe, M. (2019). A computational procedure for the recognition and classification of maize leaf diseases out of healthy leaves using convolutional neural networks. AgriEngineering, 1(1), 119-131.
38. Kim, M. (2021). Apple leaf disease classification using superpixel and CNN. In Advances in Computer Vision and Computational Biology: Proceedings from IPCV'20, HIMS'20, BIOCOMP'20, and BIOENG'20 (pp. 99-106). Cham: Springer International Publishing.
39. Khan, A. I., Quadri, S. M. K., & Banday, S. (2021). Deep learning for apple diseases: classification and identification. International Journal of Computational Intelligence Studies, 10(1), 1-12.
40. Auza-Santivañez JC, Lopez-Quispe AG, Carías A, Huanca BA, Remón AS, Condo-Gutierrez AR, et al. Improvements in functionality and quality of life after aquatic therapy in stroke survivors. AG Salud 2023;1:15-15.
41. Pushpa, B. R., Ashok, A., & AV, S. H. (2021, September). Plant disease detection and classification using deep learning model. In 2021 third international conference on inventive research in computing applications (ICIRCA) (pp. 1285-1291). IEEE.
42. Bhatt, P., Sarangi, S., Shivhare, A., Singh, D., & Pappula, S. (2019, February). Identification of Diseases in Corn Leaves using Convolutional Neural Networks and Boosting. In ICPRAM (pp. 894-899).
43. Ouchra, Hafsa, and Abdessamad Belangour. "Object Detection Approaches in Images: A Weighted Scoring Model based Comparative Study." International Journal of Advanced Computer Science and Applications 12, no. 8 (2021): 268-275.
FINANCING
The authors did not receive financing for the development of this research.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORSHIP CONTRIBUTION
Conceptualization: Khaoula Taji, Fadoua Ghanimi.
Data curation: Khaoula Taji, Fadoua Ghanimi.
Formal analysis: Khaoula Taji, Fadoua Ghanimi.
Research: Khaoula Taji, Fadoua Ghanimi.
Methodology: Khaoula Taji, Fadoua Ghanimi.
Project management: Khaoula Taji, Fadoua Ghanimi.
Resources: Khaoula Taji, Fadoua Ghanimi.
Software: Khaoula Taji, Fadoua Ghanimi.
Supervision: Khaoula Taji, Fadoua Ghanimi.
Validation: Khaoula Taji, Fadoua Ghanimi.
Display: Khaoula Taji, Fadoua Ghanimi.
Drafting - original draft: Khaoula Taji, Fadoua Ghanimi.
Writing - proofreading and editing: Khaoula Taji, Fadoua Ghanimi.