doi: 10.56294/dm202395
ORIGINAL
A detailed study on implementing new approaches in the Game of Life
Un estudio detallado sobre la aplicación de nuevos enfoques en el Juego de la Vida
1Aristotle University, 54124. Thessaloniki, Greece.
Cite as: Triantafyllou SA. A detailed study on implementing new approaches in the Game of Life. Data and Metadata 2023;2:95. https://doi.org/10.56294/dm202395.
Submitted: 10-09-2023 Revised: 05-11-2023 Accepted: 28-12-2023 Published: 29-12-2023
Editor: Prof.
Dr. Javier González Argote ![]()
Note: paper presented at the International Conference on Artificial Intelligence and Smart Environments (ICAISE’2023).
ABSTRACT
In 1952, Alan Turing who is considered as a father of Computer Science, based on his previous scientific research on the theory of computation, he emphasized how important is the analysis of pattern formation in nature and developed a theory. In his theory, he described specific patterns in nature that could be formed from basic chemical systems. Turing in his previous studies in the theory of computation, he had constantly worked on symmetrical patterns that could be formed simultaneously and realized the necessity for further analysis of pattern formation in biological problems. However, it was until the late 1960s, when John Conway was the first to introduce the “Game of Life”, an innovative mathematical game based on cellular automata, having a purpose to utilize the fundamental entities, called as cells, in two possible states described as “dead” or “alive”. This paper tries to contribute to a better understanding of the “Game of Life” by implementing algorithmic approaches of this problem in PASCAL and Python programming languages. Also, inside the paper numerous variations and extensions of the Conway’s Game of Life are proposed that introduce new ideas and concepts. Furthermore, several machine learning algorithms to learn patterns from large sets of Game of Life simulations and generate new rules or strategies are described in detail.
Keywords: Game of Life; Cellular Automata; Algorithms.
RESUMEN
En 1952, Alan Turing, considerado el padre de la informática, basándose en su investigación científica previa sobre la teoría de la computación, destacó la importancia del análisis de la formación de patrones en la naturaleza y desarrolló una teoría. En su teoría, describió patrones específicos en la naturaleza que podían formarse a partir de sistemas químicos básicos. Turing, en sus estudios previos sobre la teoría de la computación, había trabajado constantemente sobre patrones simétricos que podían formarse simultáneamente y se dio cuenta de la necesidad de profundizar en el análisis de la formación de patrones en los problemas biológicos. Sin embargo, no fue hasta finales de la década de 1960, cuando John Conway fue el primero en introducir el “Juego de la Vida”, un innovador juego matemático basado en autómatas celulares, con el propósito de utilizar las entidades fundamentales, llamadas como células, en dos posibles estados descritos como “muerto” o “vivo”. Este trabajo trata de contribuir a una mejor comprensión del “Juego de la Vida” mediante la implementación de aproximaciones algorítmicas de este problema en los lenguajes de programación PASCAL y Python. Asimismo, dentro del trabajo se proponen numerosas variaciones y extensiones del Juego de la Vida de Conway que introducen nuevas ideas y conceptos. Además, se describen en detalle varios algoritmos de aprendizaje automático para aprender patrones a partir de grandes conjuntos de simulaciones del Juego de la Vida y generar nuevas reglas o estrategias.
Palabras clave: Juego de la Vida; Autómatas Celulares; Algoritmos.
INTRODUCCIÓN
The "Game of Life" was first proposed by John Conway, and it is a cellular automaton which rules were firstly introduced by him in the late 1960s.(1,2,3,4,5,6,7,8,9,10) The detailed description of Conway’s work by Martin Gardner in the March 1970 Mathematical Games column in Scientific American, made the "Game of Life" popular to the public.(7,11) Apparently, the "living" is considered as an example of metaphor in the "Game of Life" and this mathematical game has the potential of a universal Turing machine, which means that it is constructed in a way to be computed algorithmically.(4,8,9) This Turing machine is constructed from patterns in the Conway’s "Game of Life" cellular automaton.(5,6) Conway described a method of developing a register machine which can simulate a Turing machine.(5,9) The Game of Life, invented by him is essentially a cellular automaton, where a selection of entities called as cells are arranged in a grid of an explicitly predetermined shape. Each cell gets a state from a binary set and updates its state according to a set of strict rules. Every cell transforms its state as a function of time and for Conway's "Game of Life", the cells include two states, named as "live" and "dead", and the necessary rules are driven by the states of the 8 neighboring cells, that are in the "alive" state.(1,4,7)
Τhe first cellular automaton which was developed by John von Neumann in 1966 was complex with its 29 states,(12,13,14) but there are much simpler ones which have only two states. John von Neumann’s cellular automaton was a complex automaton with 29 different states based on a Universal Turing Machine that can recreate itself. Later, Christopher G. Langton in 1984, made the things simpler when he created a cellular automaton with only eight states, which gives up the universality, but still can execute a program (Langton’s Loops) that can recreate itself.(15) A cellular automaton is a dynamic system arranged in a specified grid of cells each of which has a specific number of connected discrete states n, and in this paper, we consider n=2 (for the states set as "live" and "dead").(12,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31) Each state is updated in discrete time steps with a time function according to specific state rules.(12,14,32,33,34,35,36)
PROPOSED METHOD
Detailed Algorithm Analysis
We consider a rectangular axis system with integer subdivisions and specifically we have chosen 9 x 9 subdivisions. Every position could be empty or possessed by a "living" organism with the symbol "*". Supposing that we are in a first state called as (FIRST GENERATION), we can move to the next state by following the basic rules:
• Every position and for example the x position has 8 neighbors, the (1,2,3,4,5,6,7,8)
• A "living" organism ("*") could be extinct (death state) and its position to be possessed by a blank symbol, if and only if, there are lesser than 2 or more than 3 "living" organisms as neighbors.
• In an empty position an organism can be "born" (birth state), if and only if, in this position there are exactly 3 "living" organisms as neighbors.
Implementation of the algorithmic approaches in the PASCAL Programming Language
The decision to implement the algorithmic approaches in Pascal programming language was taken after careful consideration, because Pascal is a procedural programming language for good programming practices by using structured programming.(16,37-39) In this section we program in PASCAL programming language and the source code that is presented in detail, implements the following actions:
1. Create a rectangular axis system 9 x 9 (subroutine - procedure SCREEN).
2. Create a first state (subroutine - procedure FIRST GENERATION).
3. Create a next state starting from an existing state (subroutine - procedure NEXTGENERATION).
4. Finish the generation processes (subroutine - function FINISH).
5. With the help of the subroutines 1-4, a main program that starts from a first state moves to the creation of new generations, and terminates the overall process when needed, by using the function FINISH. The source code in PASCAL programming language is the following (see the code in PASCAL programming language):
program PROBLEM(input,output);
uses Crt;
const size = 9;
type mchar = array[0..size+1, 0..size+1] of char;
var chessboard: mchar; generation: integer;
procedure SCREEN(var chessboard: mchar; generation: integer);
var i, j: 1..size;
BEGIN
ClrScr;
WRITELN;
WRITELN('generation : ', generation);
WRITELN;
WRITE(' 1 2 3 4 5 6 7 8 9 ');
WRITELN;
FOR i := 1 TO size DO
WRITE(chessboard[i,j]:3);
WRITELN
END;
procedure FIRSTGENERATION(var chessboard: mchar; var generation: integer);
var i, j: integer; flag: boolean;
BEGIN
generation := 1;
FOR i := 0 TO size + 1 DO
FOR j := 0 TO size + 1 DO
chessboard[i,j] := ' ';
flag := FALSE;
WRITE('to terminate the process give : ');
WRITELN('( i < 1) OR ( i >', size : 2, ' OR j < 1 OR j > ', size : 2, ' ) ');
WRITELN;
REPEAT
WRITE('give the coordinates i and j of the organism : ');
READLN(i,j);
IF (i > 0) AND (i <= size) AND (j > 0) AND (j <= size)
THEN
chessboard[i,j] := '*';
ELSE
flag := TRUE
UNTIL flag
END;
procedure NEXTGENERATION(var chessboard: mchar);
var i, j: integer; helpboard: mchar; i1, j1: 0..size+1; neighbors: integer;
BEGIN
neighbors := 0;
FOR i1 := i - 1 TO i + 1 DO
FOR j1 := j - 1 TO j + 1 DO
IF chessboard[i1,j1] = '*'
THEN neighbors := neighbors + 1;
IF chessboard[i,j] = '*'
THEN neighbors := neighbors - 1;
helpboard := chessboard;
FOR i := 1 TO size DO
FOR j := 1 TO size DO
BEGIN
neighbors := 0;
FOR i1 := i - 1 TO i + 1 DO
FOR j1 := j - 1 TO j + 1 DO
IF chessboard[i1,j1] = '*'
THEN neighbors := neighbors + 1;
IF chessboard[i,j] = '*'
THEN neighbors := neighbors - 1;
IF helpboard[i,j] = ' '
THEN
BEGIN
IF neighbors = 3
THEN
helpboard[i,j] := '*';
END
ELSE IF (neighbors < 2) OR (neighbors > 3)
THEN
helpboard[i,j] := ' ';
chessboard := helpboard;
END
END;
function FINISH: boolean;
var a: char;
BEGIN
WRITE('next generation ? (for yes press: y) : ');
READLN(a);
FINISH := (a = 'y')
END;
BEGIN
ClrScr;
FIRSTGENERATION(chessboard, generation);
SCREEN(chessboard, generation);
WHILE FINISH DO
BEGIN
WRITELN;
NEXTGENERATION(chessboard);
generation := generation + 1;
SCREEN(chessboard, generation)
END
END.
Presentation of Data and Outcomes
In this section, we present the data and final outcomes after running all the subroutines and the main program in PASCAL programming language (see the following code and figure 1).
• to terminate the process give : (i < 1) OR (i > 9 OR j < 1 OR j > 9) give the coordinates i and j of the organism: 4 5
• give the coordinates i and j of the organism: 5 6
• give the coordinates i and j of the organism: 6 6
• give the coordinates i and j of the organism: 4 6
• give the coordinates i and j of the organism: 0 0
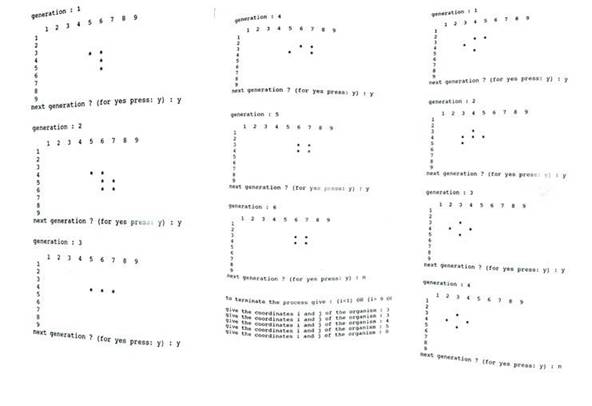
Figure 1. Data and Outcomes
Numerous variations and extensions of the Conway’s Game of Life that introduce new ideas and concepts
While the basic rules of Conway’s Game of Life are well-established, there have been numerous variations and extensions of the game that introduce new ideas and concepts.(19,22,24,25,26) Here are a few innovative approaches and ideas related to the Game of Life cellular automata:
• Higher Dimensions: exploration of the Game of Life in three or more dimensions. In a 3D grid, cells would have 26 neighbors instead of 8, leading to even more complex patterns and behaviors. Visualizing these higher-dimensional automata can be challenging but fascinating.
• Rule Variations: experimenting with different rulesets beyond Conway’s original rules leads to observe that there are countless possibilities for birth, survival, and death conditions, leading to diverse and unexpected outcomes. Some rules might promote the growth of complex structures, while others could lead to rapid decay.
• Interactive Game of Life: creation of interactive versions of the Game of Life where users can interact with the grid in real-time. They can add or remove cells, change the rules on the fly, or even influence the evolution of the patterns using various tools and controls.
• Evolutionary Algorithms: integration of evolutionary algorithms into the Game of Life to evolve patterns based on specific criteria. For example, by using genetic algorithms or other evolutionary techniques it is achievable to create populations of patterns that exhibit desired traits, such as stability, speed of movement, or complexity.
• Multiplayer Game of Life: development of a multiplayer version of the Game of Life where multiple players can influence the grid simultaneously. Players can compete or cooperate to create specific patterns or disrupt each other’s structures, leading to dynamic and competitive gameplay.
• Dynamic Rule Changes: it is possible the rules of the Game of Life to change dynamically based on the evolving patterns. For example, certain structures or arrangements could trigger rule modifications, leading to a constantly changing environment where patterns adapt and evolve in response to their surroundings.
• Visualizations and Sonifications: creation of artistic visualizations or sonifications based on the evolving patterns in the Game of Life. For instance, it is possible to translate the changing cell states into music, animations, or other forms of art, providing a sensory experience of the automaton's behavior.
• Multi-State Cellular Automata: extending the binary states of live and dead cells to a multi-state system leads every cell to have multiple states. Also, the rules can be defined to allow transitions between these states based on the states of neighboring cells. This introduces a new layer of complexity and potential for diverse patterns.
• Rule Learning and Machine Learning: exploration of machine learning techniques to discover interesting rules or predict the evolution of specific patterns. For example, it is possible to use neural networks or other machine learning algorithms to learn patterns from large sets of Game of Life simulations and generate new rules or strategies for evolving particular structures.
• Probabilistic Cellular Automata: incorporating probabilistic rules where the transition of a cell from one state to another is not deterministic but probabilistic helps to introduce randomness in the rules that can lead to stochastic patterns and behaviors, adding an element of unpredictability to the automaton.
• Community-driven Evolution: by creating an online platform where users can propose and vote on new rules or modifications to existing rules helps to allow the community to collectively influence the evolution of the Game of Life, leading to a diverse set of rulesets and patterns generated by collaborative efforts. By exploring these ideas and combining them with the fundamental principles of cellular automata, researchers and enthusiasts can continue to push the boundaries of what is possible with the Game of Life and similar automata systems.
• Interactive Evolutionary Interfaces: developing interactive interfaces that allow users to draw, modify, or manipulate patterns directly and observe how they evolve based on the rules helps to provide real-time feedback on the viability and stability of the created patterns, encouraging users to explore and experiment with different designs. These ideas describe the vast potential for exploration and creativity within the realm of cellular automata, inspiring researchers, developers, and enthusiasts to continue experimenting and pushing the boundaries of what is possible with the Game of Life and similar automata systems.
Machine learning algorithms to learn patterns from large sets of Game of Life simulations and generate new rules or strategies
Using machine learning algorithms to learn patterns from large sets of Game of Life simulations and generate new rules or strategies is an intriguing area of research.(19,22,24,25,26)
The following is an approach outlining how this could be accomplished:
• Data Collection: generate a diverse set of initial configurations for the Game of Life and simulate their evolution over many generations. Record the states of cells at each time step. These simulations will form the dataset for training the machine learning models.
• Feature Extraction: extract relevant features from the simulation data that can capture the characteristics of evolving patterns. These features could include the density of live cells, the presence of specific structures, the frequency of pattern oscillations, and other metrics describing the evolving patterns.
• Labeling the Dataset: label the dataset based on specific criteria. For instance, you might label certain patterns as "interesting" if they exhibit complex behavior, stability, or unique structures. Patterns that lead to rapid expansion or contraction could also be labeled differently. The labeling process provides supervised learning signals for the machine learning models.
• Model Selection: choose appropriate machine learning algorithms for the task. Neural networks, particularly recurrent neural networks (RNNs) or long short-term memory networks (LSTMs), can capture sequential patterns over time steps. Alternatively, you can explore other algorithms like decision trees, random forests, or even genetic algorithms, depending on the nature of the problem.
• Training the Models: train the selected machine learning models using the labeled dataset. The models learn to recognize patterns associated with specific labels. During training, the models adjust their parameters to minimize prediction errors and improve their ability to classify patterns.
• Rule Generation: once the model is trained, it can be used to generate new rules or strategies. For example, in the context of the Game of Life, the model could predict which initial configurations are likely to lead to interesting, stable, or complex patterns. These predictions can guide the generation of new rules or strategies for creating specific types of patterns.
• Evaluation and Refinement: evaluate the generated rules or strategies by simulating the Game of Life with these rules and observing the resulting patterns. Refine the machine learning models and training process based on the quality and diversity of the generated patterns. Iteratively improve the models to achieve more accurate and desirable outcomes.
• Pattern Analysis and Visualization: analyze the results to gain insights into the learned patterns. Visualize the clusters or predicted labels to understand the similarities and differences between different patterns. Visualization tools such as t-SNE or PCA can help reduce high-dimensional feature space to 2D or 3D for better understanding.
• Pattern Generation and Exploration: use the trained models to generate new patterns by providing specific input conditions or by exploring the latent space of learned patterns. Experiment with the generated patterns to see if they align with known Game of Life structures or if they reveal novel configurations.
• Iterative Exploration: continue the iterative process of generating new rules, evaluating the patterns, and refining the models. Experiment with different model architectures, features, and training strategies to explore the vast space of possible rules and strategies within the Game of Life. By following this approach, machine learning algorithms can be leveraged to discover novel patterns, rules, and strategies within the Game of Life, showcasing the potential of combining artificial intelligence with cellular automata simulations.
t-SNE (t-distributed Stochastic Neighbor Embedding) and PCA (Principal Component Analysis)
Both t-SNE (t-distributed Stochastic Neighbor Embedding) and PCA (Principal Component Analysis) are dimensionality reduction techniques commonly used for visualizing high-dimensional data in a lower-dimensional space. While these techniques are not specific to the Game of Life, they can be applied to visualize patterns and structures generated by the Game of Life simulations. An approach of utilizing t-SNE and PCA to visualize Game of Life patterns is the following:
• Data Preparation: prepare the data representing Game of Life patterns. This data should be in a structured format where each row corresponds to a pattern, and the columns represent features extracted from the patterns.
• Feature Extraction: extract relevant features from the Game of Life patterns and ensure that the features capture the important characteristics of the patterns.
• Data Normalization: normalize the feature data so that all features have similar scales. Standardizing the features (subtracting mean and dividing by standard deviation) is a common normalization technique.
• Dimensionality Reduction: a) PCA (Principal Component Analysis): apply PCA to reduce the dimensions of the feature space. PCA identifies the principal components, which are linear combinations of the original features that capture the most variance in the data. You can visualize patterns in the reduced-dimensional space using the first few principal components. An example in Python using scikit-learn is the following:
from sklearn.decomposition import PCA
# Apply PCA
pca = PCA(n_components=2)
# Reduce to 2 dimensions for visualization
reduced_data_pca = pca.fit_transform(normalized_data)
# Visualization using reduced_data_pca
# (Plotting code will depend on the plotting library you are using)
b) t-SNE (t-distributed Stochastic Neighbor Embedding): Apply t-SNE to reduce the dimensions further. t-SNE is particularly good at capturing non-linear relationships in the data. It projects the data points into a lower-dimensional space while preserving local similarities between data points. An example in Python using scikit-learn is the following:
from sklearn.manifold import TSNE
# Apply t-SNE
tsne = TSNE(n_components=2, perplexity=30, random_state=42) reduced_data_tsne = tsne.fit_transform(normalized_data)
# Visualization using reduced_data_tsne
# (Plotting code will depend on the plotting library you are using)
• Visualization: once a person has the reduced-dimensional data (either through t-SNE or PCA), he/she can use a plotting library like Matplotlib, Seaborn, or Plotly in Python to visualize the patterns in 2D. Each point in the visualization represents a Game of Life pattern, and the patterns with similar characteristics will be clustered together in the reduced-dimensional space. We should remember that the choice of parameters, such as the number of components for PCA or the perplexity for t-SNE, can significantly impact the visualization results.
CONCLUSIONS AND FUTURE WORK
The "Game of Life" cellular automaton is a characteristic paradigm of a parallel collision-based computing machine, and its rules were invented by John Conway in the late 1960s. Nowadays, we are still getting inspiration from Conway’s discovery. Dr. Conway was among one of the pioneers in the field of cellular automata and by introducing the "Game of Life", he introduced new scientific achievements in the domain of complexity science, with simulations that can be used to identify patterns and model problems like ants to traffic and many others such as clouds to galaxies. This paper tried to contribute to a better understanding of the "Game of Life" by implementing algorithmic approaches of this problem in PASCAL and Python programming languages. Also, inside the paper numerous variations and extensions of the Conway's Game of Life are proposed that introduce new ideas and concepts. Furthermore, several machine learning algorithms to learn patterns from large sets of Game of Life simulations and generate new rules or strategies are described in detail. Future studies aim to study further the "Game of Life" and its dynamics to identify patterns and model many important problems.
REFERENCES
1.Bays, C. (2010). Introduction to Cellular Automata and Conway’s Game of Life. In: Adamatzky, A. (eds) Game of Life Cellular Automata. Springer, London. https://doi.org/10.1007/978-1-84996-217-9_1
2. Caballero, L., Hodge, B. and Hernandez, S. (2016) “Conway's ‘Game of life’ and the epigenetic principle,” Frontiers in Cellular and Infection Microbiology, 6. Available at: https://doi.org/10.3389/fcimb.2016.00057.
3. Wainwright, R. (2010). Conway’s Game of Life: Early Personal Recollections. In: Adamatzky, A. (eds) Game of Life Cellular Automata. Springer, London. https://doi.org/10.1007/978-1-84996-217-9_2
4. Rendell, P. (2002). Turing Universality of the Game of Life. In: Adamatzky, A. (eds) Collision-Based Computing. Springer, London. https://doi.org/10.1007/978-1-4471-0129-1_18
5. Rendell P. Conway’s Game Life Turing Machine http://www.rendell.uk.co/gol
6. Turing A.M. On computable numbers, with applications to the entscheidungsproblem Proc. London Math. Soc. 42 (1937 230–265.
7. M. Gardner. Mathematical games: The fantastic combinations of John Conway's new solitaire Game 'Life'. Scientific American, 1970. 5, 11
8. Yurii Rogozhin. Small universal Turing machines. Theor. Comput. Sci., 168(2): 215-240, November 1996. ISSN 0304-3975. 2, 13, 46
9. Smith. Universality of Wolfram's 2, 3 Turing Machine, 2007. the Wolfram 2,3 Turing Machine Research Prize. 55, 185
10. S. Wolfram. Universality and complexity in cellular automata. Physica, 10D: 1-35, 1984. 4, 6, 13, 55, 185, 200
11. Martin Gardner, Mathematical games: The fantastic combinations of john conway’s new solitaire game “life”, Scientific American 223 (1970), 120–123.
12. Kazakov, Dimitar & Sweet, Matthew. (2004). Evolving the Game of Life. Lecture Notes in Computer Science. 3394. 10.1007/978-3-540-32274-0_9.
13. Hirte, R. (2022). John Horton Conway’s Game of Life, An overview and examples.
14. John von Neumann, Theory of self-reproducing automata, Edited by Arthur W. Burks(1966).
15. Christopher G Langton, Self-reproduction in cellular automata, Physica D: Nonlinear Phenomena 10 (1984), no. 1-2, 135–144.
16. Zaks, R. (1981). Introduction to Pascal (including UCSD Pascal). SYBEX Inc..
17. Triantafyllou, S.A. (2023). A Quantitative Research About MOOCs and EdTech Tools for Distance Learning. In: Auer, M.E., El-Seoud, S.A., Karam, O.H. (eds) Artificial Intelligence and Online Engineering. REV 2022. Lecture Notes in Networks and Systems, vol 524. Springer, Cham. https://doi.org/10.1007/978-3-031-17091-1_52
18. Triantafyllou, S.A. (2022). TPACK and Toondoo Digital Storytelling Tool Transform Teaching and Learning. In: Florez, H., Gomez, H. (eds) Applied Informatics. ICAI 2022. Communications in Computer and Information Science, vol 1643. Springer, Cham. https://doi.org/10.1007/978-3-031-19647-8_24
19. Adamatzky, A. (Ed.). (2010). Game of life cellular automata (Vol. 1). London: Springer.
20. Triantafyllou, S. A. (2022). "Work in progress: Educational Technology and Knowledge Tracing Models," 2022 IEEE World Engineering Education Conference (EDUNINE), 2022, pp. 1-4, https://doi.org/10.1109/EDUNINE53672.2022.9782335.
21. Triantafyllou, S. A. (2022). Magic squares in order 4K+2. 2022 30th National Conference with International Participation (TELECOM). https://doi.org/10.1109/TELECOM56127.2022.10017312
22. Rennard, J. P. (2002). Implementation of logical functions in the Game of Life. In Collisionbased computing (pp. 491-512). London: Springer London.
23. Triantafyllou, S.A. (2023). A Detailed Study on the 8 Queens Problem Based on Algorithmic Approaches Implemented in PASCAL Programming Language. In: Silhavy, R., Silhavy, P. (eds) Software Engineering Research in System Science. CSOC 2023. Lecture Notes in Networks and Systems, vol 722. Springer, Cham. https://doi.org/10.1007/978-3-031-35311-6_18
24. Springer, J. M., & Kenyon, G. T. (2021). It's hard for neural networks to learn the game of life. In 2021 International Joint Conference on Neural Networks (IJCNN) (pp. 1-8). IEEE.
25. Durand, B., & Róka, Z. (1999). The game of life: universality revisited. In Cellular Automata: a Parallel Model (pp. 51-74). Dordrecht: Springer Netherlands.
26. Rendell, P. (2011). A universal turing machine in conway's game of life. In 2011 International Conference on High Performance Computing & Simulation (pp. 764-772). IEEE.
27. Rodríguez-Pérez JA. Strengthening the Implementation of the One Health Approach in the Americas: Interagency Collaboration, Comprehensive Policies, and Information Exchange. Seminars in Medical Writing and Education 2022;1:11-11. https://doi.org/10.56294/mw202211.
28. Farhaoui, Y.and All, Big Data Mining and Analytics, 2023, 6(3), pp. I–II, DOI: 10.26599/BDMA.2022.9020045
29. Gonzalez-Argote D, Gonzalez-Argote J. Generation of graphs from scientific journal metadata with the OAI-PMH system. Seminars in Medical Writing and Education 2023;2:43-43. https://doi.org/10.56294/mw202343.
30. Farhaoui, Y. , "Big data analytics applied for control systems" Lecture Notes in Networks and Systems, 2018, 25, pp. 408–415. https://doi.org/10.1007/978-3-319-69137-4_36
31. Rodríguez FAR, Flores LG, Vitón-Castillo AA. Artificial intelligence and machine learning: present and future applications in health sciences. Seminars in Medical Writing and Education 2022;1:9-9. https://doi.org/10.56294/mw20229.
32. Alaoui, S.S., and all. "Hate Speech Detection Using Text Mining and Machine Learning", International Journal of Decision Support System Technology, 2022, 14(1), 80. DOI: 10.4018/IJDSST.286680
33. Alaoui, S.S.,and All "Data openness for efficient e-governance in the age of big data", International Journal of Cloud Computing, 2021, 10(5-6), pp. 522–532, https://doi.org/10.1504/IJCC.2021.120391
34. Vidal AAR de C. Modelamiento y análisis del comportamiento de la variable: Generación de Energía Eléctrica, en el sector eléctrico peruano utilizando la metodología Box and Jenkins, para la predicción de este recurso. Sincretismo 2020;1.
35. Tarik, A., and all."Recommender System for Orientation Student" Lecture Notes in Networks and Systems, 2020, 81, pp. 367–370. https://doi.org/10.1007/978-3-030-23672-4_27
36. Sossi Alaoui, S., and all. "A comparative study of the four well-known classification algorithms in data mining", Lecture Notes in Networks and Systems, 2018, 25, pp. 362–373. https://doi.org/10.1007/978-3-319-69137-4_32
FINANCING
No financing.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORSHIP CONTRIBUTION
Conceptualization: Serafeim A. Triantafyllou.
Research: Serafeim A. Triantafyllou.
Drafting - original draft: Serafeim A. Triantafyllou.
Writing - proofreading and editing: Serafeim A. Triantafyllou.