doi: 10.56294/dm202384
ORIGINAL
Extraction of fetal electrocardiogram signal based on K-means Clustering
Extracción de la señal del electrocardiograma fetal basada en el agrupamiento K-means
Mohammed Moutaib1 ![]() , Mohammed
Fattah1
, Mohammed
Fattah1 ![]() , Yousef
Farhaoui2
, Yousef
Farhaoui2 ![]() *,
Badraddine Aghoutane3
*,
Badraddine Aghoutane3 ![]() , Moulhime
El Bekkali4
, Moulhime
El Bekkali4 ![]()
1IMAGE Laboratory, Moulay Ismail University. Meknes, Morocco.
2L-STI, T-IDMS, University of Moulay Ismail, Faculty of Science and Technics. Errachidia, Morocco.
3FS, My Ismail University. Meknes, Morocco.
4IASSE Laboratory, Sidi Mohamed Ben Abdellah University. Fez, Morocco
Cite as: Moutaib M, Fattah M, Farhaoui Y, Aghoutane B, Bekkali ME. Extraction of fetal electrocardiogram signal based on K-means Clustering. Data and Metadata 2023; 2:84. https://doi.org/10.56294/dm202384.
Submitted: 03-08-2023 Revised: 19-10-2023 Accepted: 28-12-2023 Published: 29-12-2023
Editor: Prof.
Dr. Javier González Argote ![]()
Note: paper presented at the International Conference on Artificial Intelligence and Smart Environments (ICAISE’2023).
ABSTRACT
Fetal electrocardiograms (ECG) provide crucial information for the interventions and diagnoses of pregnant women at the clinical level. Maternal signals are robust, making retrieval and detection of Fetal ECGs difficult. In this article, we propose a solution based on Machine Learning by adapting the k-means clustering to detect the fetal ECG by recording the ECGs. In our first preprocessing part, we tried normalized and segmented ECG waveform. Next, we used the Euclidean distance to measure similarity. To identify a certain number of centroids in our data, the results classified into two classes are represented in the last part through graphs and compared with other algorithms, such as the CNN classifier, to demonstrate the effectiveness of this innovative approach, which can be deployed in real-time.
Keywords: ECG; Machine Learning; K-means; Fetal electrocardiograms.
RESUMEN
Los electrocardiogramas (ECG) fetales proporcionan información crucial para las intervenciones y diagnósticos de las embarazadas a nivel clínico. Las señales maternas son robustas, lo que dificulta la recuperación y detección de ECGs fetales. En este artículo, proponemos una solución basada en Machine Learning adaptando el clustering de k-means para detectar el ECG fetal mediante la grabación de los ECGs. En nuestra primera parte de preprocesamiento, intentamos normalizar y segmentar la forma de onda del ECG. A continuación, utilizamos la distancia euclídea para medir la similitud. Para identificar un cierto número de centroides en nuestros datos, los resultados clasificados en dos clases se representan en la última parte mediante gráficos y se comparan con otros algoritmos, como el clasificador CNN, para demostrar la eficacia de este enfoque innovador, que puede aplicarse en tiempo real.
Palabras clave: ECG; Machine Learning; K-means; Electrocardiogramas Fetales.
INTRODUCCIÓN
In today’s world, machine learning models are increasingly implemented worldwide for people segmentation and anomaly detection. In the field of health, the diagnostic phase is essential for the orientation of the patient and his follow-up.(1) Machine learning brings new solutions to healthcare professionals to save time and optimize the correct diagnosis. It opens new perspectives in the identification of diseases. For example, it can help doctors more easily detect abnormalities in patients’ organs or predict future diseases in the human body. Monitoring the fetus’s condition should be constant during a woman’s pregnancy to ensure good health. Continuous monitoring allows physicians to increase their presence and make better decisions faster in an emergency.
The objective is not to replace the doctor with the machine but to support him in analyzing and interpreting the enormous amount of data collection.(2,3) Machine Learning also makes it possible to promote good diagnoses and fight against medical errors by generating differential diagnoses and suggesting complementary examinations. They seem capable of solving many health problems doctors encounter, particularly interpreting ECG, EMG, and EEG biological signals.(4) Congenital heart defects are the leading cause of death from congenital disabilities. For this, one of the best techniques is monitoring the cardiac signal, which gives us essential information about the state of the fetus.(5)
The fetal electrocardiogram (ECG_Fetal) can be monitored by placing electrodes on the mother's abdomen. However, it is weak and confused with multiple noise sources, including the mother's ECG, which is unusually high in amplitude. Several approaches to extract ECG_F from signals picked up by electrodes implanted on the surface of the mother's body have been proposed in previous research.(6,7,8,9) However, these approaches have been treated with 2 methods, and the first requires many dangerous sensors for the fetus's health.(10) The second is to use another deep learning approach, such as CNN.(11)
The identification of arrhythmias has the potential to be simplified into a straightforward and practical categorization of heartbeats. As a result, numerous techniques have been proposed in this field, as referenced in several studies.(12,13) With the advancements in artificial intelligence (AI), AI has emerged as a robust solution to various challenges that previously lacked effective solutions. These challenges include classifying intricate models, predictive tasks, and computer vision.
One approach involves employing deep learning architectures within the realm of AI methods. In these architectures, the initial layers composed of convolutional neurons serve as feature extractors, while the fully connected layers at the end play a pivotal role in arriving at the final decision.(14,15)
This article focuses on one of the most powerful algorithms, K-means. It is one of the most popular clustering algorithms. It makes it possible to analyze a data set characterized by descriptors to group similar data into groups or clusters.(6,17) He proposes a new approach using a classifier based on this kind of deep architecture to classify ECG beats. The second section of this article provides our approach's methodology with a description of our database. The third section gives our proposed algorithm and the data extraction method. The last part is devoted to the result and comparing our approach with other methods to demonstrate our proposed approach's effectiveness.
SYSTEM DESCRIPTION AND METHODOLOGY
Dataset visualization
It is impractical to ask doctors to review the voluminous data collected from the human body or scans. However, the results of the processing performed on the data should be presented to the physicians in a clear and classified format where they can easily understand the interrelationships between the collected information. Visualization is considered an independent tool and an important area of research, with many applications in science and everyday life.
In this part, we are interested in visualizing the data collected on the bodies of pregnant women and the analyses. However, our application repels several problems that hinder the application in the real case, such as the cost of the material, the lack of experience in the medical field. These cited problems related to data collection guided us toward the use of authentic databases that help us visualize accurate patient data.(18)
Our research selected the direct abdominal and fetal ECG database (ADFECGDB), which can be accessed at https://physionet.org/physiobank/database/ADFECGDB, as our primary data source. These data were gathered from the Department of Obstetrics at the Medical University of Silesia using the KOMPOREL system. This system utilizes ECG (electrocardiography) sensors to capture the electrical signals generated by the heart, enabling the assessment of cardiac activity, including heart rate and the time intervals between heartbeats. Electrocardiography (ECG) represents the electrical potential that regulates heart muscle activity.(19,20) The potential is collected by electrodes placed on the skin's surface, allowing for the detection of various cardiac irregularities. ECG plays a vital role in diagnostic examinations in the field of cardiology.
Recordings were obtained from five pregnant women aged between 38 and 41 weeks. Each recording contains four differential signals (Figure 1) acquired from the maternal abdomen and a baseline direct fetal ECG obtained from the fetal head.
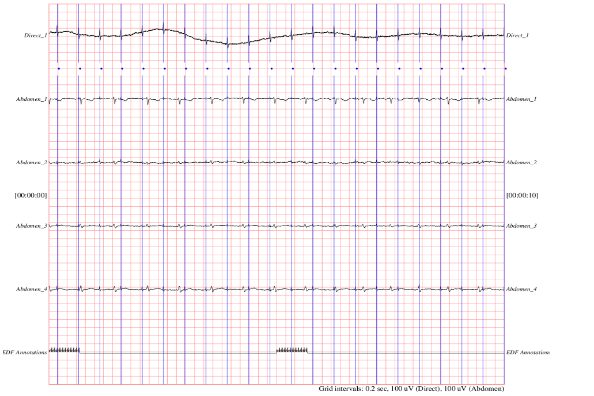
Figure 1. Representation of collected data ECG
FETAL ECG signal extraction based on the K MEANS algorithm
Our innovative approach is based on a carefully designed two-phase methodology to efficiently process and analyze data figure 2. The first phase, pre-processing, is of crucial importance as an essential prerequisite. During this stage, the raw data undergoes a process of cleaning, normalization and transformation. The goal of preprocessing is to eliminate noise and inconsistencies, ensure data consistency, and prepare it for the next phase. The clustering phase, the second step of our approach, revolves around the exploitation of preprocessed data. This step aims to group the data into homogeneous sets, thereby identifying underlying structures or patterns. Using advanced clustering techniques, our method can discover relationships, trends, and similar groups of data within the initial set. Clustering makes it much easier to interpret data by organizing it logically, which can be essential for informed decision-making and in-depth analysis.
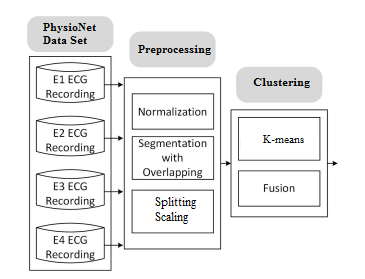
Figure 2. Data Processing Workflow
By skillfully combining preprocessing for data preparation and clustering for exploring their internal structures, our approach offers a comprehensive solution for data management and analysis. It can be successfully applied in a variety of areas, from geospatial data mining to customer segmentation, providing valuable insights for strategic decision-making and meaningful discoveries. As shown in figure 2, this is demonstrated by seeking to group the data into K distinct clusters in the observations of the data set. However, based on the data's similarity, similar data end up in the same cluster. Moreover, an observation can only be found in one cluster at a time. The same observation cannot, therefore, belong to two different clusters. An ECG sensor deployed in the mother's abdomen measures a combination of the information. In mathematical terms, our equation is interpreted as follows:
X=aFE+bMe+B (1)
knowing that Fe= ECG Fetal, Me=ECG Maternal, B =Noise.
Kmeans ECG Clustering
In order to organize our data into separate clusters, the K-Means algorithm requires a method for assessing the level of similarity between various data points. Consequently, data that closely resemble each other exhibit a smaller dissimilarity distance, whereas dissimilar objects have a greater separation distance. In our case, we chose to use the Euclidean distance, which is the geometric distance that is calculated as follows:
![]() (2)
(2)
After loading the data to separate the F and M ECGs, we must separate our data into two categories figure 3: training and test. The first category helps us train our model, and the second is used to test the model's success rate. It is the essential step of the preprocessing.

Figure 3. Data separation and Splitting
According to our algorithm, we must first define the number K of clusters. The problem is finding an optimal K. One of the most popular methods for doing this is the elbow method. The idea is to perform k-means clustering for a range of k clusters, and for each figure 4 value, we compute intraclass. When plotting the figure 5 distortions and the plot looks like an arm, the curve's inflection point is the best value of k.

Figure 4. The optimal number of cluster
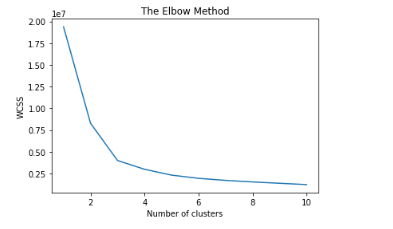
Figure 5. The Elbow Method
We notice in figure 5 that the curve is shaped like an arm. According to Elbow’s method, the optimal value of K is either two or three. This agrees with the dataset used divided into two or three classes.In our case and according to EQ 1, if we neglect noise B, we fall into two classes: the first is that of the Fetal, and the second is that of the Maternal. Our equation, therefore, is reformulated as follows:
X=aFE+bMe (3)
We are now going to implement the K-Means algorithm with two classes in figure 6:

Figure 6. Initialize the number of clusters
The result of this application is:
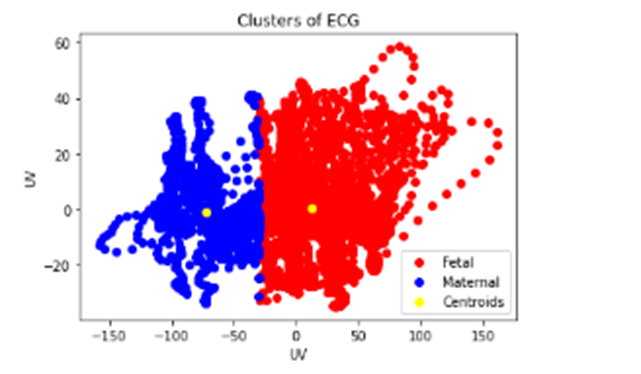
Figure 7. Results of clusters
In figure 6, we successfully deploy the K-Means algorithm, an unsupervised learning method, to classify our data into two distinct classes: fetal and maternal. This implementation is designed to solve the problem of segmenting our data based on two well-defined categories. The K-Means process begins with the initialization of two cluster centers, which serve as starting points for classification. Then, each data point is assigned to the closest cluster, based on Euclidean distance. This step is repeated iteratively until convergence, when the cluster centers no longer change significantly. The result of this implementation is the division of our data into two clusters figure 7 shows us two classes of observations in red and blue, respectively, which describe the excellent separation of fetal and maternal data, each grouping together data points sharing similar characteristics.
RESULTS AND DISCUSSION
To verify the performance of our proposed K-means algorithm, we performed simulations in the following 5-axis ECG recording. In figure 8, we present the results of the simulation based on the approach described in this work to determine the MECG and FECG signals. According to the global algorithm, we start by building the images of our data separated from axis 1, which demonstrates the variance of the Maternal ECG, and the same for axis 2, which presents the ECG of the Fetal. In Axis 3, we presented the 2 ECGs to compare their changes. Due to its low amplitude, the contribution of the fetus in terms of energy is meager. However, the powerful fetal heartbeat allows it to be easily illustrated in the time-frequency and time-scale domains, which is the main focus of this article. In axes 4 and 5, the Fetal and Maternal ECGs in summer are present with their RR interval.
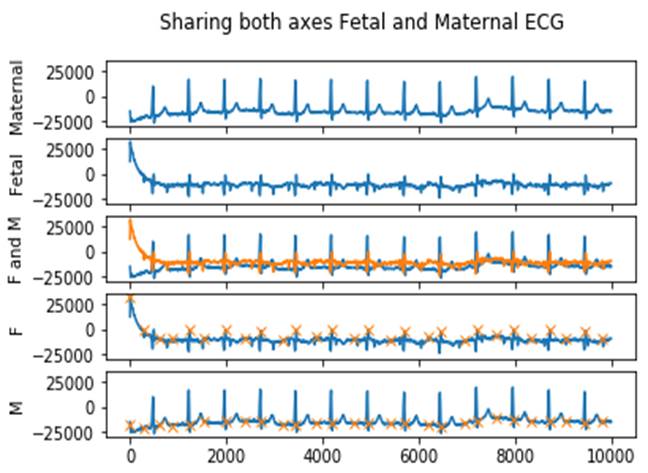
Figure 8. Sharing both axes, Fetal and Maternal ECG
Comparison with Related Work
So far, several methods of extracting MECG and FECG signals have been carried out. Among these methods,(21) have proposed an empirical mode decomposition method(22,23) and have proposed a least error algorithm mean squares. A solution that combines 3 algorithms, ICA, TS, and EKF, has been proposed.(24) They used the TS subtraction algorithm and the ICA algorithm to extract the Fetal and Maternal signals and then EKF to filter the Fetal and Maternal ECG components. One of the main requirements of(25) is to improve the 'FastICA' algorithm by adding an element to deal with the initial weight in the iterative Newton algorithm, which reduces the iteration time. Compared to previous work,(26) sought to improve the number of cross-relations under the same number of iterations. Figure 1 compares a set of proposed algorithms on works with their performance.
Similarly, proposed a method to estimate Fetal and Maternal ECG signals using the EKS approach with an adaptive system to estimate the actual ECG component. The results showed that our proposed Kmeans algorithm had higher accuracy.(27) The K-Means algorithm identifies several centroids in a data set, a centroid being the arithmetic mean of all data points belonging to a particular cluster. In recent years, several types of research have been conducted applying deep learning algorithms, such as several studies,(28,29,30,31,32,33) which have sought to process the signals. Studies have identified the characteristics of FECG signals using neural networks.(34,35,36,37,38,39) Figure 9 indicates that when extracting R waves, the efficiency and performance of the algorithms are similar to our proposed algorithm. However, deep learning algorithms demonstrate efficiency and performance in extracting the characteristics of low-amplitude signals because they can select a specific signal automatically. Studies have an approach based on convolutional CNNs as a solution that automatically extracts the signal and determines the fetal ECG.(40,41,42,43,44) Compared to our proposed algorithm, the disadvantage of these approaches is that the generalizability and accuracy of the model could not simply be increased only if adding more samples would impact the model's accuracy. On the other hand, our algorithm is deployed to discover groups that have not been explicitly defined and classify a dataset into several groups according to our approach, regardless of the size of our database.(45,46,47)
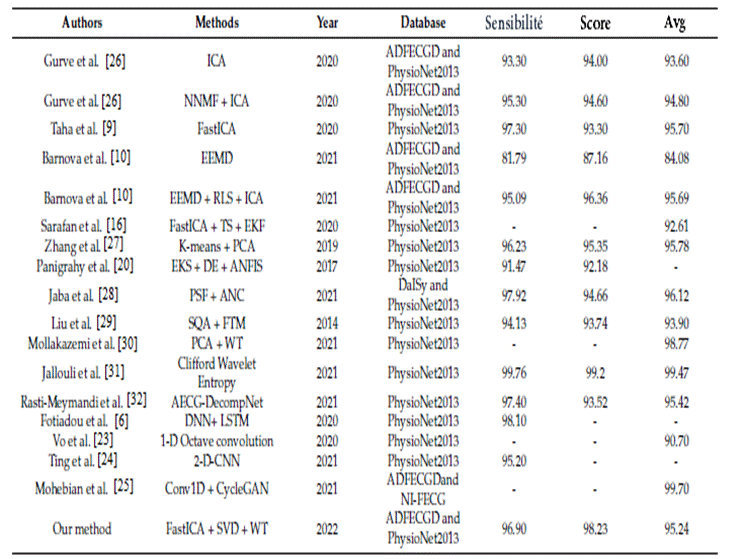
Figure 9. Comparison of methods used in related works for the extraction of FECG and MECG signals
* Score: is the positive predictive value, and Avg is the harmonic mean of precision and recall
CONCLUSION
Fetal electrocardiograms have become a way to recognize fetal heart disease and change. However, extracting this signal is very important and challenging in medical practice. In this article, we have proposed a K-means Clustering algorithm to obtain Fetal and Maternal signal separates. This algorithm has been integrated into a graphical interface. The proposed method can be used as a new Fetal signal extraction and detection method. In future work, deep learning methods and multi-channel signals can be considered.
REFERENCES
1. Moutaib, M., Ahajjam, T., Fattah, M., Farhaoui, Y., Aghoutane, B., & el Bekkali, M. (2022). Reduce the Energy Consumption of IOTs in the Medical Field. Digital Technologies and Applications, 259 268. https://doi.org/10.1007/978-3-031-02447-4_27.
2. Moutaib, M., Ahajjam, T., Fattah, M., Farhaoui, Y., & Aghoutane, B. (2021). Reduce the Energy Consumption of Connected Objects. Proceedings of the 2nd International Conference on Big Data, Modelling and Machine Learning. https://doi.org/10.5220/0010728900003101
3. M. Moutaib, M. Fattah, Y. Farhaoui, Internet of things: Energy Consumption and Data Storage, Procedia Computer Science, Volume 175, 2020, Pages 609-614.
4. Monson, M.; Heuser, C.; Einerson, B.D.; Esplin, I.; Snow, G.; Varner, M.; Esplin, M.S. Evaluation of an external fetal electrocardiogram monitoring system: A randomized controlled trial. Am. J. Obstet. Gynecol. 2020, 223, e1–e244.
5. Zwanenburg, F.; Jongbloed, M.R.M.; Van Geloven, N.; Ten Harkel, A.D.J.; Van Lith, JMM; Haak, M.C. Assessment of human fetal cardiac autonomic nervous system development using color tissue Doppler imaging. Echocardiography 2021, 38, 974–981.
6. Fotiadou, E.; Xu, M.; Van Erp, B.; Van Sloun, R.J.G.; Vullings, R. Deep Convolutional Long Short-Term Memory Network for Fetal Heart Rate Extraction. In Proceedings of the 42nd Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Montreal, QC, Canada, 20–24 July 2020; pp. 608–611.
7. M. Moutaib, T. Ahajjam, M. Fattah, Y. Farhaoui, B. Aghoutane, and M. el Bekkali (2021). Optimization of the Energy Consumption of Connected Objects. International Journal of Interactive Mobile Technologies (iJIM), 15(24), 176 190. https://doi.org/10.3991/ijim.v15i24.26985
8. Sulas, E.; Urru, M.; Tumbarello, R.; Raffo, L.; Pani, D. Systematic analysis of single-and multi-reference adaptive filters for non-invasive fetal electrocardiography. Math. Biosci. Eng. 2019, 17, 286–308.
9. Taha, L.; Abdel-Raheem, E. A null space-based blind source separation for fetal electrocardiogram signals. Sensors 2020, 20, 3536.
10. Barnova, K.; Martinek, R.; Jaros, R.; Kahankova, R.; Matonia, A.; Jezewski, M.; Czabanski, R.; Horoba, K.; Jezewski, J. A novel algorithm based on ensemble empirical mode decomposition for non-invasive fetal ECG extraction. PLoS ONE 2021, 16, e0256154
11. Wu, S.; Shen, Y.; Zhou, Z.; Lin, L.; Zeng, Y.; Gao, X. Research of fetal ECG extraction using wavelet analysis and adaptive filtering.Comput. Biol. Med. 2013, 43, 1622–1627.
12. Vasudeva, B.; Deora, P.; Pradhan, P.M.; Dasgupta, S. Efficient implementation of LMS adaptive filter-based FECG extraction on an FPGA. Healthc. Technol. Lett. 2020, 7, 125–131.
13. Ferranti, M.; Le, T.H.; Vandebril, R. A comparison between the complex symmetric based and classical computation of the singular value decomposition of normal matrices. Numer. Algorithms 2021, 67, 109–120.
14. Kumar, A.; Tomar, H.; Mehla, V.K.; Komaragiri, R.; Kumar, M. Stationary wavelet transform based ECG signal denoising method.ISA Trans. 2021, 114, 251–262.
15. Martinek, R.; Kahankova, R.; Jezewski, J.; Jaros, R.; Mohylova, J.; Fajkus, M.; Nedoma, J.; Janku, P.; Nazeran, H. Comparative effectiveness of ICA and PCA in extraction of fetal ECG from abdominal signals: Toward non-invasive fetal monitoring. Front. Physiol. 2018, 9, 648.
16. Sarafan, S.; Le, T.; Naderi, A.M.; Nguyen, Q.D.; Kuo, B.T.Y.; Ghirmai, T.; Han, H.D.; Lau, M.P.H.; Cao, H. Investigation of methods to extract fetal electrocardiogram from the mother's abdominal signal in practical scenarios. Technologies 2020, 8, 33.
17. Hyvärinen, A. Fast and robust fixed-point algorithms for independent component analysis. IEEE Trans. Neural Netw. 1999, 10, 626–634.
18. Yuan, L.; Zhou, Z.; Yuan, Y.;Wu, S. An improved FastICA method for fetal ECG extraction. Comput. Math. Methods Med. 2018,2018, 7061456
19. Kaleem, A.M.; Kokate, R.D. A survey on FECG extraction using neural network and adaptive filter. Soft Comput. 2021, 25,4379–4392.
20. Panigrahy, D.; Sahu, PK Extraction of fetal ECG signal by an improved method using extended Kalman smoother framework from single channel abdominal ECG signal. Australas. Phys. Eng. Sci. Med. 2017, 40, 191–207.
21. Vijayakumar, V.; Ummar, S.; Varghese, T.J.; Shibu, A.E. ECG noise classification using deep learning with feature extraction.Signal. Image Video P. 2022, 1–7.
22. Tseng, K.K.; Wang, C.; Xiao, T.J.; Chen, C.M.; Hassan, M.M.; Albuquerque, V.H.C. Sliding large kernel of deep learning algorithm for mobile electrocardiogram diagnosis. Comput. Electr. Eng. 2021, 96, 107521
23. Vo, K.; Le, T.; Rahmani, A.M.; Dutt, N.; Cao, H. An efficient and robust deep learning method with 1-D octave convolution to extract fetal electrocardiogram. Sensors 2020, 20, 3757.
24. Ting, Y.C.; Lo, F.W.; Tsai, P.Y. Implementation for fetal ECG detection from multi-channel abdominal recordings with 2D convolutional neural network. J. Signal. Process. Syst. 2021, 93, 1101–1113.
25. Mohebbian, M.R.; Vedaei, S.S.;Wahid, K.A.; Dinh, A.; Marateb, H.R.; Tavakolian, K. Fetal ECG extraction from maternal ECG using attention-based CycleGAN. IEEE. J. Biomed. Health 2022, 26, 515–526.
26. Gurve, D.; Krishnan, S. Separation of fetal-ECG from single-channel abdominal ECG using activation scaled non-negative matrix factorization. IEEE. J. Biomed. Health Inform. 2020, 24, 669–680.
27. Zhang, Y.; Yu, S. Single-lead noninvasive fetal ECG extraction by means of combining clustering and principal components analysis. Med. Biol. Eng. Comput. 2020, 58, 419–432.
28. Jaba, D.K.A.; Dhanalakshmi, S.R.K. An improved parallel sub-filter adaptive noise canceler for the extraction of fetal ECG. Biomed. Tech. 2021, 66, 503–514.
29. Liu, C.; Li, P.; Di, MC; Zhao, L.; Zhang, H.; Chen, Z. A multi-step method with signal quality assessment and fine-tuning procedure to locate maternal and fetal QRS complexes from abdominal ECG recordings. Physiol. Meas. 2014, 35, 1665–1683.
30. Mollakazemi, M.J.; Asadi, F.; Tajnesaei, M.; Ghaffari, A. Fetal QRS Detection in Noninvasive Abdominal Electrocardiograms Using Principal Component Analysis and DiscreteWavelet Transforms with Signal Quality Estimation. J. Biomed. Phys. Eng. 2021,11, 197–204.
31. Chafi, Saad-Eddine, et al. “Resource placement strategy optimization for smart grid application using 5G wireless networks.” International Journal of Electrical and Computer Engineering, Volume 12, Issue 4, Pages 3932 – 3942, 2022. https://doi.org/10.11591/ijece.v12i4.pp3932-3942
32. Chafi, Saad-Eddine, et al. “Cloud computing services, models and simulation tools.” International Journal of Cloud Computing, vol. 10, no. 5–6, pp. 533–547, 2021. https://doi.org/10.1504/IJCC.2021.120392
33. D. Daghouj, M. Fattah, S. Mazer, Y. Balboul, and M. El Bekkali, “UWB waveform for automotive short range radar,” International Journal on Engineering Applications, vol. 8, no. 4, pp. 158–164, Jul. 2020. https://doi.org/10.15866/irea.v8i4.18997
34. M. Abdellaoui, M. Fattah, “Characterization of Ultra Wide Band indoor propagation In 7th Mediterranean Congress of Telecommunications (CMT). IEEE, 2019, https://doi.org/10.1109/CMT.2019.8931367
35. Mamane, M. Fattah, M. el Ghazi, Y. Balboul, M. el Bekkali, and S. Mazer, “The impact of scheduling algorithms for real-time traffic in the 5G femto-cells network,” 9th International Symposium on Signal, Image, Video and Communications, ISIVC 2018 , Pages 147 – 1512, July 2018, https://doi.org/10.1109/ISIVC.2018.8709175
36. Farhaoui, Y., “Design and implementation of an intrusion prevention system” International Journal of Network Security, vol.19(5), pp. 675–683, 2017. DOI: 10.6633/IJNS.201709.19(5).04
37. Farhaoui, Y.and All, Big Data Mining and Analytics, 2023, 6(3), pp. I–II, DOI: 10.26599/BDMA.2022.9020045
38. Farhaoui, Y., “Intrusion prevention system inspired immune systems” Indonesian Journal of Electrical Engineering and Computer Science, vol. 2(1), pp. 168–179, 2016.
39. Farhaoui, Y. , "Big data analytics applied for control systems" Lecture Notes in Networks and Systems, 2018, 25, pp. 408–415. https://doi.org/10.1007/978-3-319-69137-4_36
40. Farhaoui, Y. and All, Big Data Mining and Analytics, 2022, 5(4), pp. I IIDOI: 10.26599/BDMA.2022.9020004
41. Alaoui, S.S., and all. "Hate Speech Detection Using Text Mining and Machine Learning", International Journal of Decision Support System Technology, 2022, 14(1), 80. DOI: 10.4018/IJDSST.286680
42. Alaoui, S.S., and all. ,"Data openness for efficient e-governance in the age of big data", International Journal of Cloud Computing, 2021, 10(5-6), pp. 522–532, https://doi.org/10.1504/IJCC.2021.120391
43. El Mouatasim, A., and all. "Nesterov Step Reduced Gradient Algorithm for Convex Programming Problems", Lecture Notes in Networks and Systems, 2020, 81, pp. 140–148. https://doi.org/10.1007/978-3-030-23672-4_11
44. Tarik, A., and all."Recommender System for Orientation Student" Lecture Notes in Networks and Systems, 2020, 81, pp. 367–370. https://doi.org/10.1007/978-3-030-23672-4_27
45. Sossi Alaoui, S., and all. "A comparative study of the four well-known classification algorithms in data mining", Lecture Notes in Networks and Systems, 2018, 25, pp. 362–373. https://doi.org/10.1007/978-3-319-69137-4_32
46. Farhaoui, Y."Teaching Computer Sciences in Morocco: An Overview", IT Professional, 2017, 19(4), pp. 12–15, 8012307. DOI: 10.1109/MITP.2017.3051325
47. Farhaoui, Y., "Securing a Local Area Network by IDPS Open Source", Procedia Computer Science, 2017, 110, pp. 416–421. https://doi.org/10.1016/j.procs.2017.06.106
FINANCING
The authors did not receive financing for the development of this research.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORSHIP CONTRIBUTION
Conceptualization: Mohammed Moutaib, Mohammed Fattah, Yousef Farhaoui, Badraddine Aghoutane, Moulhime El Bekkali.
Research: Mohammed Moutaib, Mohammed Fattah, Yousef Farhaoui, Badraddine Aghoutane, Moulhime El Bekkali.
Drafting - original draft: Mohammed Moutaib, Mohammed Fattah, Yousef Farhaoui, Badraddine Aghoutane, Moulhime El Bekkali.
Writing - proofreading and editing: Mohammed Moutaib, Mohammed Fattah, Yousef Farhaoui, Badraddine Aghoutane, Moulhime El Bekkali.