doi: 10.56294/dm2024453
SHORT COMMUNICATION
Worldwide genetic variability of the rs1861868 SNP in the FTO gene associated with obesity
Variabilidad genética global del polimorfismo rs1861868 del gen FTO asociado con la obesidad
Sergio V. Flores1 ![]() *, Ángel Roco-Videla2
*, Ángel Roco-Videla2 ![]() *, Joel Antonio Herrera-Soto3
*, Joel Antonio Herrera-Soto3 ![]() , Marcela
Caviedes-Olmos4
, Marcela
Caviedes-Olmos4 ![]() *, Román M. Montaña5
*, Román M. Montaña5 ![]() *
*
1Universidad Arturo Prat. Santiago, Chile.
2Universidad Católica Silva Henríquez. Facultad de Ciencias de la Salud. Santiago, Chile.
3Universidad Bernardo O´Higgins. Dirección de Investigación, Vicerrectoría Académica, Santiago, Chile.
4Universidad de las Américas. Facultad de Salud y Ciencias Sociales. Santiago, Chile.
5Universidad Autónoma de Chile, Facultad de Ciencias de la Salud. Santiago, Chile.
Cite as: Flores SV, Roco-Videla Ángel, Herrera-Soto JA, Caviedes-Olmos M, Montaña RM. Worldwide genetic variability of the rs1861868 SNP in the FTO gene associated with obesity. Data and Metadata. 2024; 3:453. https://doi.org/10.56294/dm2024453
Submitted: 20-01-2024 Revised: 01-05-2024 Accepted: 30-07-2024 Published: 31-07-2024
Editor: Adrián
Alejandro Vitón-Castillo ![]()
ABSTRACT
Introduction: genetic predisposition to obesity is linked to an imbalance between food intake and energy expenditure, regulated by the nervous and endocrine systems. The FTO gene variants significantly impact obesity susceptibility in different populations. The objective of the research was to analyze the genetic variability of the SNP rs1861868 in the FTO gene and its association with obesity in various populations.
Method: genotype data from 1000 Genomes and allele frequencies from ALFRED were analyzed. Moran’s I assessed spatial autocorrelation, Hardy-Weinberg equilibrium was tested using VCFtools, and ANOVA compared risk allele frequencies across continents.
Results: Moran’s I indicated no significant spatial autocorrelation globally, but higher concentrations of the risk allele were observed in Europe. ANOVA showed significant differences in risk allele frequencies among continents, with Europe having the highest frequency. Hardy-Weinberg equilibrium was observed within macro populations but not globally.
Conclusions: regional variations significantly impact the distribution of the rs1861868 (T) risk allele. Evolutionary, historical, and demographic are candidate factors that shaped the genetic landscape of the FTO gene related to obesity.
Keywords: FTO; Obesity; Genetic Variability; rs1861868.
RESUMEN
Introducción: la predisposición genética a la obesidad se asocia con un desequilibrio entre la ingesta de alimentos y el gasto energético, regulado por los sistemas nervioso y endocrino Las variantes del gen FTO impactan significativamente en la susceptibilidad a la obesidad en diferentes poblaciones. El objetivo de la investigación fue analizar la variabilidad genética del SNP rs1861868 en el gen FTO y su asociación con la obesidad en diversas poblaciones.
Método: se analizaron datos de genotipos de 1000 Genomas y frecuencias alélicas de ALFRED. La autocorrelación espacial se evaluó con Moran’s I, el equilibrio de Hardy-Weinberg se probó con VCFtools y se compararon las frecuencias alélicas mediante ANOVA.
Resultados: Moran’s I no indicó una autocorrelación espacial significativa a nivel global, aunque se observó una mayor concentración del alelo de riesgo en Europa. El ANOVA mostró diferencias significativas en las frecuencias alélicas entre continentes, siendo Europa la de mayor frecuencia. Se observó equilibrio de Hardy-Weinberg dentro de las macro poblaciones, pero no a nivel global.
Conclusiones: las variaciones regionales impactan significativamente en la distribución del alelo rs1861868. Factores evolutivos, históricos y demográficos moldean el paisaje genético del gen FTO relacionado con la obesidad.
Palabras clave: FTO; Obesidad; Variabilidad Genética; rs1861868.
INTRODUCTION
The genetic predisposition to developing overweight or obesity is associated with an imbalance between food intake and energy expenditure, regulated by the nervous and endocrine systems.(1) These conditions are related to various metabolic diseases, with genetic and environmental factors influencing the increase in body mass index (BMI), which has a relatively high heritability.(2,3) Genetic markers influencing hypothalamic nuclei, regulators of appetite and satiety, or related to metabolic hormones have been identified, including the FTO gene (Fat mass and Obesity associated gene). Major polymorphisms within this gene include MC4R, ABCA1, and ADIPOQ, associated with increased body mass and the development of type 2 diabetes.(4,5)
The FTO gene was one of the first to be linked to obesity. Its main function is associated with the regulation of energy homeostasis, although specific mechanisms are not yet fully understood. Introns 1 and 2 of the FTO gene show the strongest associations with BMI and fat mass, followed by intron 3. (6) Variants in the FTO locus have significant genetic effects on obesity susceptibility in different populations.(7) Single nucleotide polymorphisms (SNPs) in the first intron of the FTO gene have been associated with obesity in various populations.(8,9,10)
Among the SNPs in the first intron, rs9939609 has been widely studied. One study found that children and adolescents with the A allele of rs9939609 had loss of control over eating high-fat foods.(11) The rs1861868, located in the same linkage disequilibrium block, has shown similar effects.(6)
The objective of the research was to analyze the genetic variability of the SNP rs1861868 in the FTO gene and its association with obesity in various populations. Understanding the distribution and frequency of this polymorphism in different population groups is important to identify genetic patterns that may contribute to obesity predisposition. Additionally, this knowledge can provide insights into the underlying biological mechanisms and help develop personalized strategies for obesity prevention and treatment.
METHOD
Genotypes from the 1000 Genomes Phase 3 database for rs1861868 were downloaded. The sample includes macro populations from Africa (N=661), East Asia (N=504), South Asia (N=489), Europe (N=503), and Latin America (N=347), covering 26 populations and a total of 2504 individuals. Additionally, allele frequencies were obtained from the ALFRED database, including 109 populations grouped into nine macro populations: Africa (N=2681), East Asia (N=1785), Central Asia (N=354), West Asia (N=292), North Asia (N=428), South Asia (N=1276), Europe (N=5494), Oceania (N=72), and Latin America (N=910). The allele frequencies of rs1861868 T were analyzed for 110 populations, totaling 13292 individuals.
Spatial autocorrelation analysis was performed using Moran’s I statistic to evaluate the spatial distribution of the risk allele globally and by continents: North America, Latin America, Africa, Europe, East Asia, and South Asia. To visualize the spatial distribution, a global frequency map was generated.
The Hardy-Weinberg equilibrium (HWE) for the 1000 Genomes data was evaluated globally and within each macro population (Africans, East Asians, South Asians, Europeans, and Latin Americans) using VCFtools. The normality of the risk allele frequency was assessed with the Kolmogorov-Smirnov test. An analysis of variance (ANOVA) was conducted to compare the mean risk allele frequencies between continents, followed by a post hoc analysis using the Tukey HSD test to identify significant differences between specific groups.
All statistical analyses were performed using R software, including Moran’s I calculation with the “spdep” package, global map generation with “worldmap,” normality tests, ANOVA, and post hoc analysis. Additionally, VCFtools was used for Hardy-Weinberg equilibrium analysis in the 1000 Genomes dataset.
RESULTS
The Kolmogorov-Smirnov test for normality did not reject the null hypothesis that the data come from a normal distribution (p= 0,27).
The results indicated a Moran’s I correlation value of -0,0092, with a Z-score of -1,0092. These results suggest no significant spatial autocorrelation in the risk allele frequency. Although overall results do not indicate significant spatial autocorrelation, a notable trend was observed in Europe, where the risk allele frequencies appear more concentrated. This concentration of high frequencies in Europe contrasts with the more uniform distribution observed in other continents, such as Asia and Africa.
To evaluate regional differences in greater detail, separate Moran’s I analyses were performed for North America, South America, Africa, Europe, East Asia, and South Asia. The results showed no significant spatial autocorrelation in any of the studied regions (figure 1).
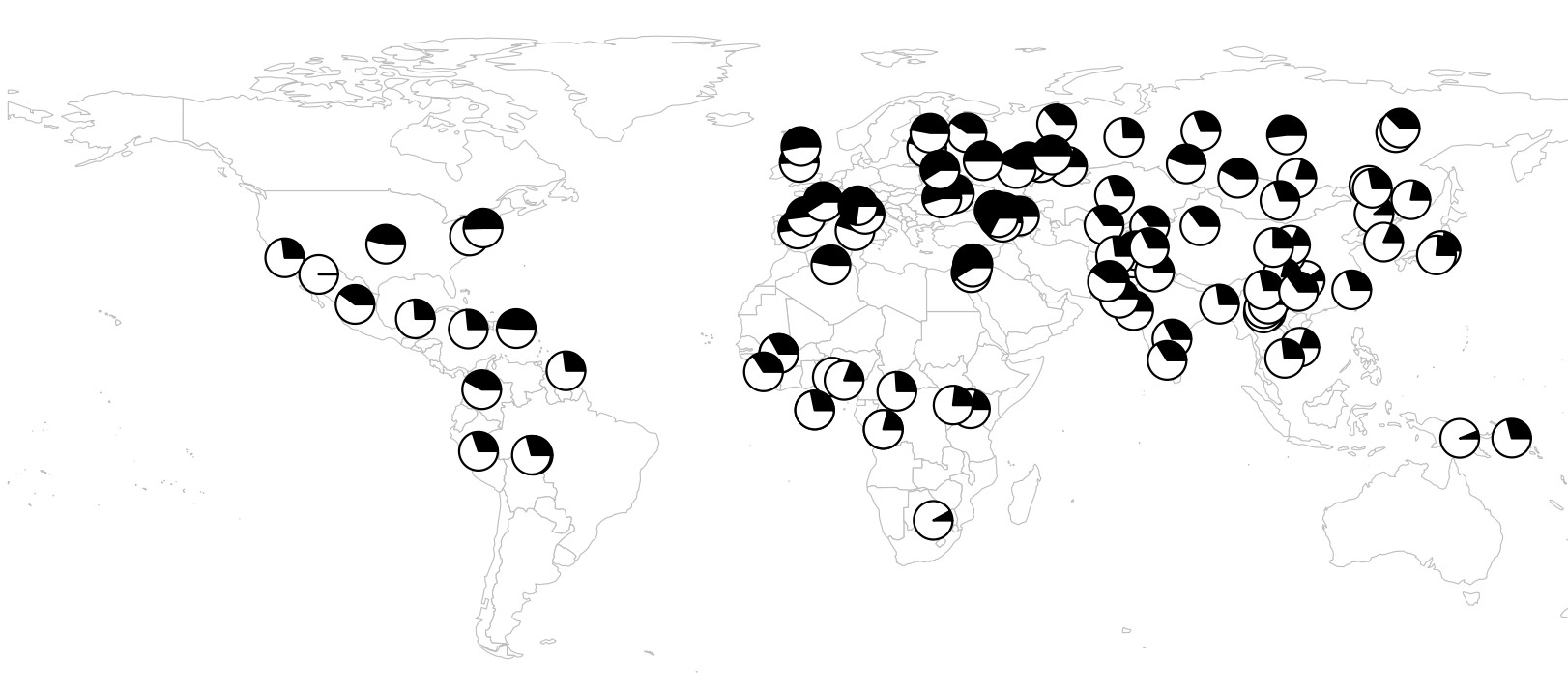
Figure 1. World map distribution of the risk allele rs1861868 (A) in the FTO gene
The ANOVA analysis to compare the risk allele frequency between North America, Latin America, Africa, Europe, East Asia, and South Asia indicated an F-statistic of 18,8665 and a p-value of 8,58 x 10-13. Table 1 shows the results of the ANOVA analysis.
|
Table 1. ANOVA results from the comparison of risk allele frequency by continents |
||||
|
Source |
Sum of Squares |
df |
F |
PR(>F) |
|
Geographical regions |
1,118195 |
6 |
18,866495 |
8,582632e-13 |
|
Residual |
0,671714 |
68 |
|
|
The results of the Tukey HSD test showed significant differences between Africa and Europe, East Asia and Europe, and between Europe and other regions (North America, South America, and South Asia). In contrast, no significant differences were observed between other pairs of regions. Europe had a significantly higher mean frequency of the risk allele (0,315) compared to Africa (0,035), East Asia (0,030), North America (0,106), South America (0,079), and South Asia (0,104). Departure from the Hardy-Weinberg equilibrium was obtained when the test was applied to the pooled populations from the 1000 Genomes dataset (p= 1,59 x 10-3), supporting the hypothesis of heterozygous deficit (p= 9,23 x 10-4). When analyzed separately, all five macro populations were in Hardy-Weinberg equilibrium (p> 0,05).
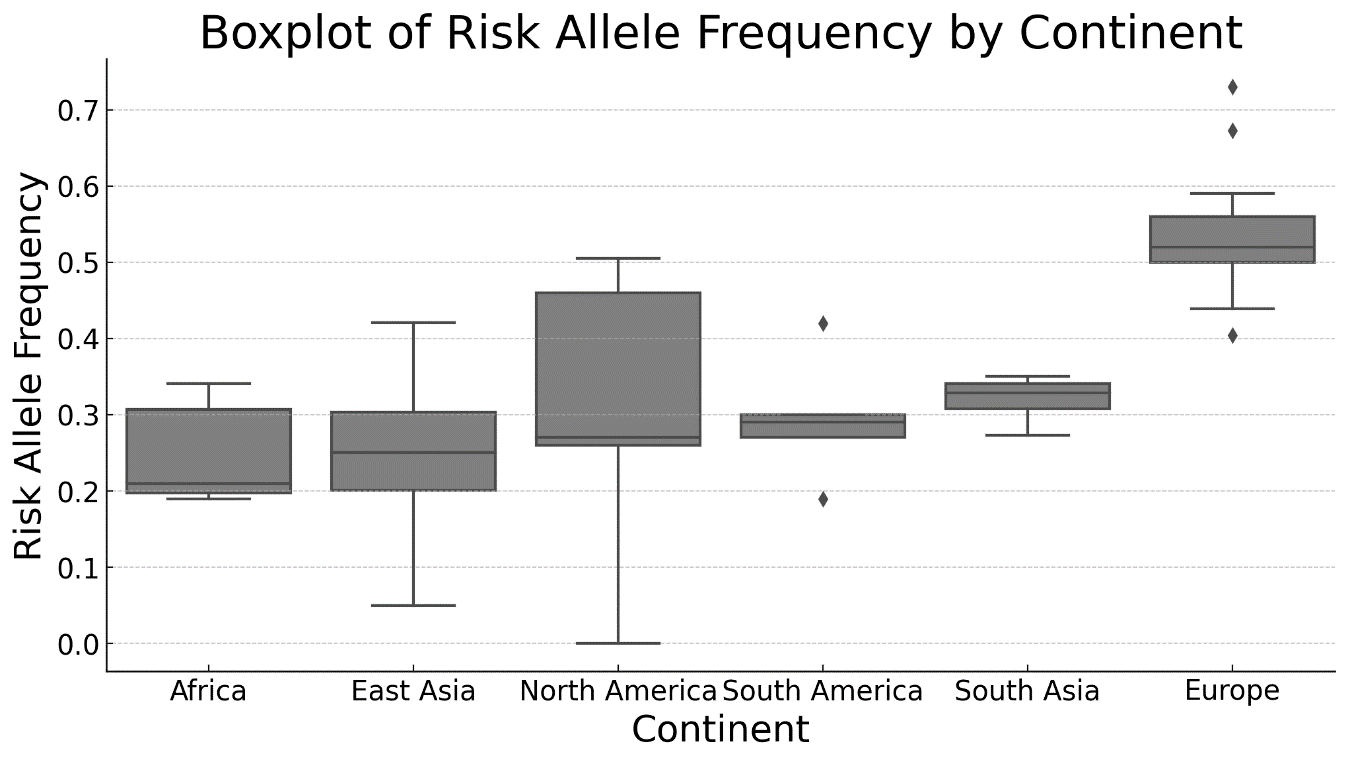
Figure 2. Boxplot of risk allele frequency rs1861868 (T) in the FTO gene in different geographic regions
DISCUSSION
The results of this study provide a comprehensive view of the distribution of the risk allele frequency rs1861868 (T) of the FTO gene, associated with obesity, in various global populations. The lack of significant spatial autocorrelation suggests that genetic variability within continents and different historical and demographic influences have affected the distribution of this allele. Although no significant spatial autocorrelation was observed globally, notable regional differences were identified, particularly in Europe, where risk allele frequencies were significantly higher compared to other regions, confirming trends in previous studies.(7,6)
The analysis of variance revealed significant differences in the risk allele frequency among continents. Europe had a significantly higher mean frequency of the risk allele compared to Africa, Asia, North America, and South America. This suggests that specific evolutionary and demographic factors may have influenced the higher prevalence of the risk allele in the European population. However, the specific biochemical mechanism of action of the FTO gene is complex, making a specific evolutionary hypothesis difficult.(4,8)
The Hardy-Weinberg disequilibrium at the global level, but not within macro populations, indicates that deviations may result from population structure and subpopulation admixture, which is relevant to consider in meta-analyses such as Liu et al. (9) The high frequency of the allele in Europe could be related to historical events of migration and population mixing, leading to the genetic homogeneity observed in this region.(10)
CONCLUSIONS
The findings suggest that evolutionary, historical, and demographic forces have shaped the distribution of the rs1861868 (T) allele of the FTO gene in different populations.
Future studies should investigate these factors further to better understand the genetic landscape of the FTO gene and its relationship with obesity.
REFERENCES
1. Gadde KM, Martin CK, Berthoud HR, Heymsfield SB. Obesity: pathophysiology and management. J Am Coll Cardiol. 2018;71(1):69-84. https://doi.org/10.1016/j.jacc.2017.11.011
2. Dobson R, Burgess MI, Sprung VS, et al. Metabolically healthy and unhealthy obesity: differential effects on myocardial function according to metabolic syndrome, rather than obesity. Int J Obes (Lond). 2016;40(1):153-161. https://doi.org/10.1038/ijo.2015.151
3. Pasqua T, Cerra MC, Angelone T. Mechanisms and pathophysiology of obesity: upgrading a complex scenario. Curr Med Chem. 2020;27(2):172-173. https://doi.org/10.2174/092986732702200218123007
4. Fawcett KA, Barroso I. The genetics of obesity: FTO leads the way. Trends Genet. 2010;26(6):266-274. https://doi.org/10.1016/j.tig.2010.02.006
5. Zayani N, Omezzine A, Boumaiza I, et al. Association of ADIPOQ, leptin, LEPR, and resistin polymorphisms with obesity parameters in Hammam Sousse Sahloul Heart Study. J Clin Lab Anal. 2017;31(6). https://doi.org/10.1002/jcla.22148
6. Tönjes A, Zeggini E, Kovacs P, Böttcher Y, Schleinitz D, Dietrich K, et al. Association of FTO variants with BMI and fat mass in the self-contained population of Sorbs in Germany. Eur J Hum Genet. 2010;18(1):104-110. https://doi.org/10.1038/ejhg.2009.107
7. Villalobos-Comparán M, Flores-Dorantes MT, Villarreal-Molina MT, Rodríguez-Cruz M, García-Ulloa AC, Robles L, et al. The FTO gene is associated with adulthood obesity in the Mexican population. Obesity. 2008;16(10):2296-2301. https://doi.org/10.1038/oby.2008.367
8. Zavattari P, Loche A, Pilia S, Ibba A, Moi L, Guzzetti C, et al. Rs9939609 in the FTO gene is associated with obesity but not with several biochemical parameters in Sardinian obese children: FTO in obese Sardinian children. Ann Hum Genet. 2011;75(6):648-654. https://doi.org/10.1111/j.1469-1809.2011.00674.x
9. Liu Y, Liu Z, Song Y, Zhou D, Zhang D, Zhao T, et al. Meta-analysis added power to identify variants in FTO associated with type 2 diabetes and obesity in the Asian population. Obesity. 2010;18(8):1619-1624. https://doi.org/10.1038/oby.2009.469
10. Song Y, You NC, Hsu YH, Howard BV, Langer RD, Manson JE, et al. FTO polymorphisms are associated with obesity but not diabetes risk in postmenopausal women. Obesity. 2008;16(11):2472-2480. https://doi.org/10.1038/oby.2008.408
11. Tanofsky-Kraff M, Han JC, Anandalingam K, Shomaker LB, Columbo KM, Wolkoff LE, et al. The FTO gene rs9939609 obesity-risk allele and loss of control over eating. Am J Clin Nutr. 2009;90(6):1483-1488. https://doi.org/10.3945/ajcn.2009.28439
FINANCING
This research was funded by the “Regular Research Project Competition”, grant number “DI-04/23”, entitled “Prevalence of rs1861868-FTO and rs7975232-VDR polymorphisms in Chilean women and their association with BMI, anthropometry and cardiovascular risk factors. according to the consumption of estrogen-based contraceptives”, awarded by the Research Department of the University of the Americas, Providencia, Chile.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORSHIP CONTRIBUTION:
Conceptualization: Sergio V. Flores.
Data curation: Sergio V. Flores, Román Montaña, Joel Antonio Herrera-Soto.
Formal analysis: Sergio V. Flores, Angel Roco-Videla.
Research: Sergio V. Flores, Angel Roco-Videla, Román Montaña.
Methodology: Sergio V. Flores, Angel Roco-Videla.
Software: Sergio V. Flores, Marcela Caviedes-Olmos.
Supervision: Sergio V. Flores, Joel Antonio Herrera-Soto.
Validation: Angel Roco-Videla, Joel Antonio Herrera-Soto.
Display: Román Montaña, Marcela Caviedes-Olmos.
Drafting - original draft: Sergio V. Flores, Angel Roco-Videla.
Writing - proofreading and editing: Angel Roco-Videla, Marcela Caviedes-Olmos.