doi: 10.56294/dm2024.354
SHORT COMMUNICATION
Association of the rs4988235(C) Polymorphism, a Determinant of Lactose Intolerance, with Genetic Ancestry in Latin American Populations
Asociación del polimorfismo rs4988235(c), determinante de intolerancia a la lactosa, y la ancestría genética en poblaciones latinoamericanas
Sergio V. Flores1
![]() *, Román M.
Montaña2
*, Román M.
Montaña2 ![]() *, Angel
Roco-Videla3
*, Angel
Roco-Videla3 ![]() *, Marcela
Caviedes-Olmos4
*, Marcela
Caviedes-Olmos4 ![]() *
*
1Universidad Arturo Prat. Santiago, Chile.
2Universidad Autónoma de Chile, Facultad de Ciencias de la Salud. Santiago, Chile.
3Universidad Bernardo O´Higgins, Programa de Magister en ciencias químico-Biológicas. Santiago, Chile.
4Universidad de las Américas. Facultad de Salud y Ciencias Sociales. Santiago, Chile.
Cite as: Flores SV, Montaña RM, Roco-Videla A, Caviedes-Olmos M. Association of the rs4988235(C) Polymorphism, a Determinant of Lactose Intolerance, with Genetic Ancestry in Latin American Populations. Data and Metadata. 2024;3:.354. https://doi.org/10.56294/dm2024.354
Submitted: 19-01-2024 Revised: 03-05-2024 Accepted: 20-09-2024 Published: 21-09-2024
Editor: Adrián
Alejandro Vitón Castillo ![]()
Corresponding author: Sergio V. Flores1 *
ABSTRACT
Introduction: the rs4988235(C) polymorphism is associated with lactose intolerance and exhibits heterogeneity among populations. In Europe, the T allele (lactose tolerance) is prevalent in the north, while the C allele (lactose intolerance) is common in Asia and Africa.
Methods: genotypes for rs4988235 were obtained from the 1000 Genomes Project database, selecting Latin American samples (Colombians, Mexican Americans, Peruvians, and Puerto Ricans). A total of 446 ancestry-informative markers (AIMs) were used to estimate genetic ancestry proportions. Shapiro-Wilks tests were conducted, and due to non-normality, non-parametric Kruskal-Wallis and post hoc Wilcoxon tests were applied.
Results: the Shapiro-Wilks test indicated significant deviations from normality for Native-American (statistic=0,8787, p<0,05) and European ancestry proportions (statistic=0,9653, p<0,05). Kruskal-Wallis analysis showed significant differences in European (statistic=26,6696, p=1,62×10−6) and Native-American (statistic=13,4306, p=0,0012) ancestry proportions among genotypes. Post hoc Wilcoxon tests indicated significant differences between Intolerant (GG) and Heterozygous (GA) genotypes for both ancestries.
Conclusions: the proportions of European and Native-American ancestry vary among genotypes of the rs4988235(C) polymorphism, suggesting the effect of admixture on the distribution of lactose intolerance in Latin American populations.
Keywords: Lactose Intolerance; rs4988235 Polymorphism; Genetic Ancestry; Latin American Populations; European Ancestry; Native-American Ancestry.
RESUMEN
Introducción: el polimorfismo rs4988235(C) está asociado con la intolerancia a la lactosa y muestra heterogeneidad entre poblaciones. En Europa, el alelo T (tolerancia a la lactosa) es prevalente en el norte, mientras que el alelo C (intolerancia) es común en Asia y África.
Métodos: se obtuvieron genotipos para rs4988235 de la base de datos del consorcio 1000 Genomas, seleccionando muestras latinoamericanas (colombianos, mexicoamericanos, peruanos y puertorriqueños). Se utilizaron 446 SNPs de marcadores informativos de ancestría para estimar las proporciones de ancestría genética. Se realizaron pruebas de Shapiro-Wilks y, debido a la no normalidad, se aplicaron pruebas no paramétricas de Kruskal-Wallis y pruebas post hoc de Wilcoxon.
Resultados: la prueba de Shapiro-Wilks indicó desviaciones significativas de la normalidad para las proporciones de nativos americanos (estadístico=0,8787, p<0,05) y de ascendencia europea (estadístico=0,9653, p<0,05). El análisis de Kruskal-Wallis mostró diferencias significativas en las proporciones de ascendencia europea (estadístico=26,6696, p=1,62×10-6) y nativa americana (estadístico=13,4306, p=0,0012) entre genotipos. Las pruebas post hoc de Wilcoxon indicaron diferencias significativas entre los genotipos Intolerante (GG) y Heterocigoto (GA) para ambas ascendencias.
Conclusiones: las proporciones de ascendencia europea y nativa americana varían entre los genotipos del polimorfismo rs4988235(C), lo que sugiere el efecto de la mezcla en la distribución de la intolerancia a la lactosa en poblaciones latinoamericanas.
Palabras clave: Intolerancia a la Lactosa; Polimorfismo Rs4988235; Ascendencia Genética; Poblaciones Latinoamericanas; Ascendencia Europea; Ascendencia Nativa Americana.
INTRODUCTION
The rs4988235(C) polymorphism, associated with lactose intolerance, exhibits heterogeneity among human populations. This SNP in the regulatory region of the MCM6 gene influences the expression of the LCT gene, responsible for the production of lactase, necessary for lactose digestion.(1) In Europe, the T allele (lactose tolerance) is prevalent in the north, while the C allele (lactose intolerance) is more common in Asia and Africa.(2,3)
The frequency of the C allele varies significantly between continents and subpopulations. In the Americas, this heterogeneity is observed with persistence frequencies varying according to genetic admixture and migration history.(4,5) Allele frequencies show high heterogeneity among ancient populations due to the first migration out of Africa around 100,000 years ago.(6) In the Americas, populated less than 20,000 years ago, this heterogeneity is due to historical admixture dynamics, suggesting the integration of genetic ancestry into the analysis of risk alleles.(7)
Genetic ancestry refers to the “architecture of genomic variation among populations”, and the individual genetic ancestry proportions result from microevolutionary processes such as panmixia, reproductive stratification, migration, and natural selection, influenced by biological and sociocultural phenomena.(8,9) Evidence of genetic ancestry can be obtained from allele configurations using specific computational methods and tools.(10) Genetic structuring, or the non-random distribution of genetic variations, is influenced by genetic drift, natural selection, migration, and non-random mating, especially in recently admixed populations like those in Latin America.(11)
This study analyzes the association between European and Native-American genetic ancestry and the rs4988235(C) allele in five Latin American populations. The hypothesis is that the tolerance allele is associated with European ancestry, while the intolerance allele is associated with Native-American ancestry.
METHODS
Genotyping for rs4988235 and Genetic Ancestry Estimation
Genotypes for rs4988235 were obtained using VcfTools(12) from the 1000 Genomes Project Consortium database.(13) From this database, only the Latin American sample was selected for further analyses, including four mixed-ancestry populations (table 1): Colombians, Mexican Americans, Peruvians, and Puerto Ricans.
|
Table 1. Population and sample size |
|
|
Population |
No. |
|
Colombians from Medellin, Colombia |
94 |
|
Mexicanamericans from Los Angeles, USA |
64 |
|
Peruvians from Lima, Peru |
85 |
|
Puerto Ricans from Puerto Rico |
104 |
|
Total Latin America |
347 |
To estimate the proportions of genetic ancestry, 446 SNPs from a panel of ancestry informative markers (AIMs) suggested by Galanter et al. were used.(14) These markers were designed, optimized, and validated to estimate genetic ancestry proportions in individuals and populations across Latin America. For this estimation, the five macro-populations included in the 1000 Genomes database were considered: African, East Asian, South Asian, European, and Latin American.
Statistical Analysis
Five ancestral populations (K=5) were modeled to estimate the genetic ancestry proportions of everyone using STRUCTURE.(15) The Shapiro-Wilks test was then performed to assess the normality of the distribution of individual genetic ancestry proportions. Since none of the ancestries followed a normal distribution (p-valor, Shapiro-Wilks < 0,05), non-parametric tests were applied: Kruskal-Wallis and post hoc Wilcoxon pairwise tests, to test the null hypothesis of no association between genetic ancestry and the presence of the lactose intolerance allele.
RESULTS
The normality analysis of Native-American and European ancestry proportions was performed using the Shapiro-Wilk test. For Native-American ancestry, the statistic was 0,8787 and the p-value was 6,63×10−16; for European ancestry, the statistic was 0,9653 and p= 2,40×10−7, both indicating significant deviations from normality (p < 0,05). The proportions of Native-American ancestry were concentrated at lower values, while those of European ancestry showed greater dispersion and a tendency toward higher values (Figure 1). These results suggest the need to use non-parametric tests to evaluate the associations between genetic ancestry and rs4988235 genotypes.
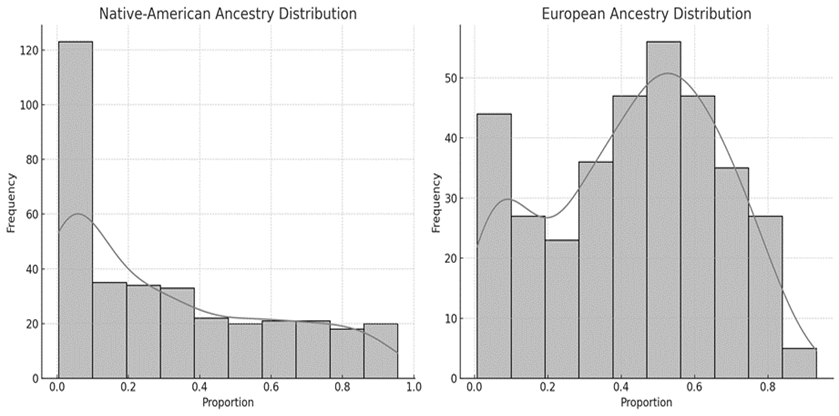
Figure 1. Distribution of Native-American ancestry and European ancestry proportions among individuals
In the Kruskal-Wallis analysis, European and Native-American ancestry proportions were compared among the genotypes of the rs4988235 polymorphism, where the G allele is associated with lactose intolerance and the A allele with tolerance. For European ancestry, the statistic was 26,6696 and the p-value was 1,62×10−6, indicating significant differences among the Intolerant (GG), Heterozygous (GA), and Tolerant (AA) genotypes. For Native-American ancestry, the statistic was 13,4306 and the p-value was 0,0012, also indicating significant differences (figure 2).
Post hoc pairwise Wilcoxon tests were conducted to compare European and Native-American ancestry proportions among the genotypes of the rs4988235 polymorphism. For European ancestry, significant differences were found between the Intolerant (GG) and Heterozygous (GA) genotypes (statistic: 8263,5, p: 0,000026) and between the Intolerant and Tolerant (AA) genotypes (statistic: 1288,5, p: 0,000327). The comparison between Heterozygous and Tolerant genotypes was not significant (statistic: 890,5, p: 0,082406). For Native-American ancestry, only the comparison between Intolerant and Heterozygous genotypes was significant (statistic: 14314,0, p: 0,000654). These results indicate significant differences in genetic ancestry proportions among certain genotypes, highlighting the importance of considering genetic ancestry in lactose intolerance studies in Latin American populations.

Figure 2. Distribution of European ancestry (left) and Native-American ancestry (right) among the genotypes of the rs4988235(C) polymorphism
DISCUSSION
The results of this study highlight the relevance of genetic ancestry in the variability of rs4988235(C) genotypes, associated with lactose intolerance, in Latin American populations. Kruskal-Wallis analyses and post hoc pairwise Wilcoxon tests revealed significant differences in European and Native-American ancestry proportions among different genotypes, suggesting that genetic admixture in these populations influences the distribution of the lactose intolerance allele.
The high frequency of the C allele, associated with lactose intolerance, in non-European populations, and its significant variability within Latin American populations, reflects the complexity of admixture and migration history in these regions. The proportion of European ancestry is higher in individuals with the A allele, which confers lactose tolerance, consistent with patterns observed in Europe, where lactase persistence is common due to historical selective pressure from dairy consumption. (2,3)
On the other hand, significant differences in Native-American ancestry proportions between the Intolerant (GG) and Heterozygous (GA) genotypes suggest that the genetics of indigenous populations also play a role in the prevalence of lactose intolerance in Latin America. This finding underscores the importance of considering both evolutionary history and sociocultural factors in studying genetic patterns in admixed populations. (9)
This study has some limitations. The sample is limited to four Latin American populations, which may not represent the full genetic diversity of the region. The use of data from the 1000 Genomes Consortium may introduce biases due to population selection. The estimation of genetic ancestry is based on a panel of SNPs that may not capture all relevant variations. Additionally, non-parametric statistical methods, while suitable for non-normal data, may have lower statistical power compared to parametric methods.
CONCLUSIONS
The proportions of European and Native-American ancestry vary significantly among the genotypes of the rs4988235(C) polymorphism, highlighting the importance of admixture in the distribution of lactose intolerance in contemporary Latin American populations.
The differences in ancestry proportions between the Intolerant (GG) and Heterozygous (GA) genotypes suggest that evolutionary history and sociocultural factors influence the population patterns of lactose intolerance.
This study demonstrates that genetic ancestry is a relevant dimension in epidemiological studies and the development of personalized medicine strategies in Latin America.
REFERENCES
1. Enattah NS, Sahi T, Savilahti E, Terwilliger JD, Peltonen L, Järvelä I. Identification of a variant associated with adult-type hypolactasia. Nat Genet. 2002;30(2):233-237. https://doi.org/10.1038/ng826
2. Itan Y, Powell A, Beaumont MA, Burger J, Thomas MG. The Origins of Lactase Persistence in Europe. PLoS Comput Biol. 2009;5(8). https://doi.org/10.1371/journal.pcbi.1000491
3. Ségurel L, Bon C. On the Evolution of Lactase Persistence in Humans. Annu Rev Genomics Hum Genet. 2017;18:297-319. https://doi.org/10.1146/annurev-genom-091416-035340
4. Jones BL, Raga TO, Liebert A, Zmarz P, Bekele E, Danielsen ET, Olsen AK, Bradman N, Troelsen JT, Swallow DM. Diversity of Lactase Persistence Alleles in Ethiopia: Signature of a Soft Selective Sweep. Am J Hum Genet. 2013;93(3):538-544. https://doi.org/10.1016/j.ajhg.2013.06.015
5. Anguita-Ruiz A, Aguilera CM, Gil Á. Genetics of Lactose Intolerance: An Updated Review and Online Interactive World Maps of Phenotype and Genotype Frequencies. Nutrients. 2020;12(9):2689. https://doi.org/10.3390/nu12092689
6. López S, Van Dorp L, Hellenthal G. Human dispersal out of Africa: a lasting debate. Evol Bioinform Online. 2015;11. https://doi.org/10.4137/EBO.S33489
7. Adhikari K, Mendoza-Revilla J, Chacón-Duque JC, Fuentes-Guajardo M, Ruiz-Linares A. Admixture in Latin America. Curr Opin Genet Dev. 2016;41:106-114. https://doi.org/10.1016/j.gde.2016.09.003
8. Dries DL. Genetic Ancestry, Population Admixture, and the Genetic Epidemiology of Complex Disease Editorial. Circ Cardiovasc Genet. 2009;2:540-543. https://doi.org/10.1161/CIRCGENETICS.109.922898
9. Creanza N, Kolodny O, Feldman MW. Cultural evolutionary theory: How culture evolves and why it matters. Proc Natl Acad Sci U S A. 2017;114(30):7782-7789. https://doi.org/10.1073/pnas.1620732114
10. Royal CD, Novembre J, Fullerton SM, Goldstein DB, Long JC, Bamshad MJ, Clark AG. Inferring genetic ancestry: opportunities, challenges, and implications. Am J Hum Genet. 2010;86(5):661-673. https://doi.org/10.1016/j.ajhg.2010.03.011
11. Chakraborty R. Analysis of Genetic Structure of Populations: Meaning, Methods, and Implications. In: Majumder PP, editor. Human Population Genetics. Springer; 1993. p. 191-224. https://doi.org/10.1007/978-1-4615-2970-5_14
12. Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, et al. The variant call format and VCFtools. Bioinformatics. 2011;27(15):2156-2158. https://doi.org/10.1093/bioinformatics/btr330
13. 1000 Genomes Project Consortium. A global reference for human genetic variation. Nature. 2015;526(7571):68. https://doi.org/10.1038/nature15393
14. Galanter JM, Fernandez-Lopez JC, Gignoux CR, Barnholtz-Sloan J, Fernandez-Rozadilla C, Via M, et al. Development of a panel of genome-wide ancestry informative markers to study admixture throughout the Americas. PLoS Genet. 2012;8(3). https://doi.org/10.1371/journal.pgen.1002554
15. Hubisz MJ, Falush D, Stephens M, Pritchard JK. Inferring weak population structure with the assistance of sample group information. Mol Ecol Resour. 2009;9(5):1322-1332. https://doi.org/10.1111/j.1755-0998.2009.02591.x
FINANCING
The authors did not receive financing for the development of this research.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORSHIP CONTRIBUTION
Conceptualization: Sergio V. Flores.
Data curation: Sergio V. Flores, Román Montaña.
Formal analysis: Sergio V. Flores, Angel Roco-Videla.
Research: Sergio V. Flores, Angel Roco, Román Montaña, Marcela Caviedes-Olmos.
Methodology: Sergio V. Flores, Angel Roco-Videla.
Software: Sergio V. Flores.
Supervision: Sergio V. Flores.
Validation: Angel Roco-Videla, Marcela Caviedes-Olmos.
Display: Román Montaña.
Drafting - original draft: Sergio V. Flores.
Writing - proofreading and editing: Angel Roco-Videla, Marcela Caviedes-Olmos.