doi: 10.56294/dm2024.379
SHORT COMMUNICATION
Genetic Native American ancestry is associated with a low likelihood of VDR rs7975232 risk genotypes for vitamin D levels
La ancestría genética nativo americana se asocia con una baja probabilidad de genotipos de riesgo del VDR rs7975232 para los niveles de vitamina D
Sergio V. Flores1 ![]() *, Angel Roco-Videla2
*, Angel Roco-Videla2 ![]() *, Román M. Montaña3
*, Román M. Montaña3 ![]() *, Marcela
Caviedes-Olmos4
*, Marcela
Caviedes-Olmos4 ![]() *, Sofia Pérez-Jiménez4
*, Sofia Pérez-Jiménez4 ![]() *, Raúl Aguilera Eguía5
*, Raúl Aguilera Eguía5 ![]() *
*
1Universidad Arturo Prat. Santiago, Chile.
2Universidad Católica Silva Henríquez, Facultad de Ciencias de la Salud. Santiago, Chile.
3Universidad Autónoma de Chile, Facultad de Ciencias de la Salud. Santiago, Chile.
4Universidad de las Américas. Facultad de Salud y Ciencias Sociales, Santiago, Chile.
5Universidad Católica de la Santísima Concepción, Departamento de Salud Pública. Concepción, Chile.
Cite as: Flores SV, Roco-Videla A, Montaña RM, Caviedes-Olmos M, Pérez-Jiménez S, Aguilera Eguía R. Genetic Native American ancestry is associated with a low likelihood of VDR rs7975232 risk genotypes for vitamin D levels. Data and Metadata. 2024; 3:.379. https://doi.org/10.56294/dm2024.379
Submitted: 23-02-2024 Revised: 12-05-2024 Accepted: 17-08-2024 Published: 18-08-2024
Editor: Adrián
Alejandro Vitón-Castillo ![]()
Corresponding autor: Sofia Pérez-Jiménez *
ABSTRACT
Introduction: obesity is associated with chronic diseases, with women being more prone, possibly due to the relationship between the α-estrogen receptor and vitamin D receptors.
Objective: the objective of this research is to analyze the distribution of VDR rs7975232 (ApaI) genotypes in Latin American populations and its relationship with genetic ancestry.
Method: 446 SNPs from an AIMs panel were used to estimate genetic ancestry proportions in individuals from Peru, Mexico, Colombia, and Puerto Rico using STRUCTURE software. Kruskal-Wallis, Mann-Whitney tests and logistic regression were applied for analyses.
Results: risk genotypes AA and CA show a low proportion of Native American ancestry and a high proportion of European and African ancestry. Logistic regression indicated an inverse effect of Native American ancestry on risk genotypes.
Conclusion: the results suggest that Native American ancestry decreases the likelihood of carrying VDR rs7975232 risk genotypes. These findings contribute to a better understanding of genetic variability and its relationship with health conditions in these populations.
Keywords: rs7975232; Genetic Ancestry; Latin American Populations; Vitamin D Receptor.
RESUMEN
Introducción: la obesidad se asocia con enfermedades crónicas, siendo las mujeres más propensas, posiblemente debido a la relación entre el receptor α de estrógeno y los receptores de vitamina D.
Objetivo: el objetivo de esta investigación es analizar la distribución de los genotipos VDR rs7975232 (ApaI) en poblaciones latinoamericanas y su relación con las ancestrías genéticas.
Método: se utilizaron 446 SNPs de un panel AIMs para estimar las proporciones de ancestría genética en individuos de Perú, México, Colombia y Puerto Rico utilizando el software STRUCTURE. Se aplicaron las pruebas de Kruskal-Wallis, Mann-Whitney y regresión logística para los análisis.
Resultados: los genotipos de riesgo AA y CA muestran una baja proporción de ancestría nativa americana y una alta proporción de ancestría europea y africana. La regresión logística indicó un efecto inverso de la ancestría nativa americana sobre los genotipos de riesgo.
Conclusión: los resultados sugieren que la ancestría nativa americana disminuye la probabilidad de portar genotipos de riesgo VDR rs7975232. Estos hallazgos contribuyen a una mejor comprensión de la variabilidad genética y su relación con las condiciones de salud en estas poblaciones.
Palabras clave: rs7975232; Ancestría Genética; Poblaciones Latinoamericanas; Receptor de Vitamina D.
INTRODUCTION
Obesity is associated with chronic diseases such as diabetes mellitus, cardiovascular diseases, neurological deficiencies and cancer, which means that obese people have a lower life expectancy.(1) On the other hand, women have a higher prevalence of obesity than men, particularly morbid obesity, which makes them a group of greater interest in terms of studying the causes of increased body mass index (BMI).(2) This greater risk of obesity in the female population may be due to the relationship between the α-estrogen receptor and the expression of vitamin D receptors, which could imply a greater deficiency, which has been associated with an increase in the obesity risk.(3,4)
The VDR (vitamin D receptor) gene regulates vitamin D levels in the human body and codes for a receptor that binds to the active form of vitamin D, 1,25-dihydroxyvitamin D3.(5,6,7) Among the most studied polymorphisms of the VDR gene is rs7975232 (ApaI), whose CA genotype has been associated with a decrease in vitamin D levels.(8) The AA genotype of rs7975232 has been associated with the risk of osteoporosis in Egyptian women, but not the CA genotype.(9)
Regarding the association of ApaI with the development of obesity and metabolic syndrome, the results are still contradictory, while in studies carried out in populations such as Thai and Iranian, the AA and CA genotypes have been associated with the development of both conditions, on the other hand. On the other hand, studies carried out in the Egyptian female population and in Turkish children have not shown that this association exists.(6,7,9,10)
The objective of this research is to analyze the population distribution of the AA and CA risk genotypes based on information from the 1000 genomes database, analyzing the influence of migratory flows on the American population and the potential risk of developing obesity. Metabolic syndrome and osteoporosis in the Latin American female population.
METHOD
Genotypes (N= 694) were downloaded from the 1000 Genomes Project database for Latin American populations: Peru, Mexico, Colombia, and Puerto Rico. To estimate the proportions of genetic ancestry, 446 SNPs from an ancestry informative markers (AIMs) panel suggested by Galanter et al. were used.(11) These markers were designed, optimized, and validated to estimate the proportions of genetic ancestry in individuals and populations across Latin America.
For this estimation, the five macro populations included in the 1000 Genomes Project database were considered: African, East Asian, South Asian, European, and Latin American. Five ancestral populations (K=5) were modeled to estimate the proportions of genetic ancestry for each individual using the STRUCTURE software.(12)
The normality of the ancestry proportions was tested using the Kolmogorov-Smirnov test. Since normality was rejected (p < 0,001), non-parametric tests were used for subsequent analyses. The contributions of Native American, European, and African genetic ancestry proportions for each of the three genotypes (AA, CA, CC) were compared using the Kruskal-Wallis test. For post hoc analyses, the Mann-Whitney test was employed.
Additionally, logistic model analyses were conducted for each of the three ancestries separately and together to evaluate the effect of these on the probability of presenting risk genotypes in individuals.
RESULTS
No Hardy-Weinberg equilibrium deviation was observed (Chi2 test, P = 0,101) for the rs7975232 genotypes, so the following analyses were continued with the entire sample. In the populations of Peru, Mexico, Colombia, and Puerto Rico, the frequencies of the risk genotypes AA and CA are presented in figure 1.
Normality was rejected for the three ancestries using the Kolmogorov-Smirnov test (p < 0,001). Due to this, non-parametric tests were used for subsequent analyses, such as Kruskal-Wallis and post hoc Mann-Whitney tests.
When analyzing the proportion of Native American ancestry in the three genotypes, it was observed that the risk genotypes AA and CA have a low proportion of this ancestry (p-value = 4,34 x 10-11). The inverse pattern was observed when analyzing the European population (p-value = 1,17 x 10-6) and African ancestry (Kruskal-Wallis p-value = 3,67 x 10-6). The post hoc analyses resulted in all comparisons being significantly different for the ancestry median (Mann-Whitney test, p < 0,01) (figure 2).
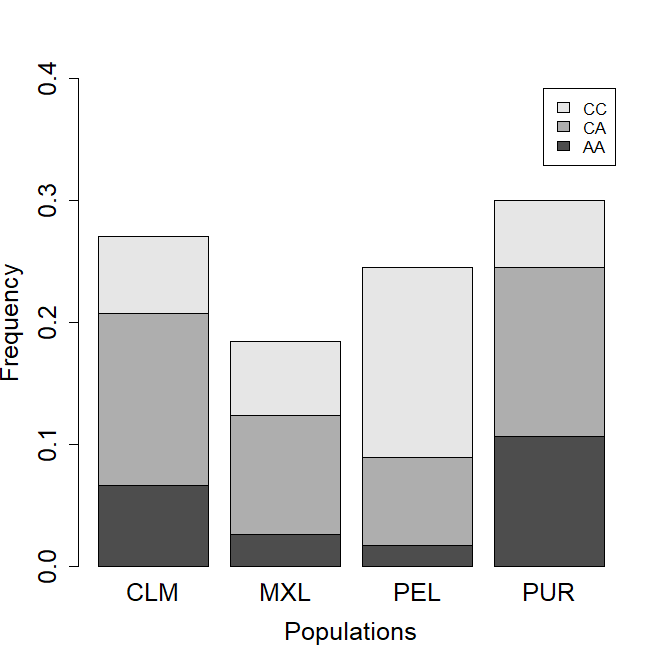
Figure 1. VDR rs7975232 genotypes frequencies in the populations here analyzed.
Note: (CLM: Colombia, MXL: México, PEL: Perú, PUR: Puerto Rico).
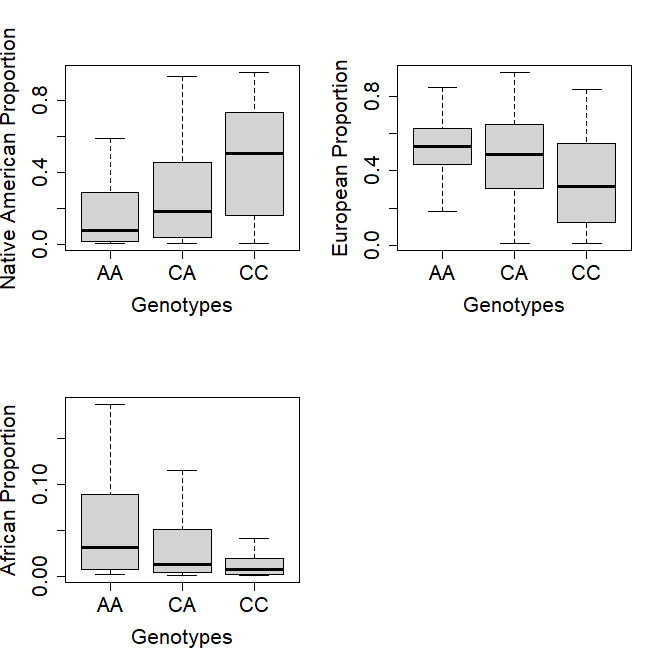
Figure 2. Genetic ancestry proportions for each VDR rs7975232 genotypes
Logistic regression through GLM resulted in an inverse effect of the proportion of Native American ancestry with a coefficient= -2,7832 and a p-value= 5,7 x 10-11. Conversely, a higher proportion of European ancestry led to a greater accumulation of risk genotypes (coefficient = 2,59, p-value= 5,71 x 10-07), as well as African ancestry (coefficient= 4,4039, p-value= 0,027445) (table 1).
|
Table 1. Results for the logistic regression model combining the genetic ancestries |
||||
|
|
Estimate |
Standard error |
Z value |
Pr(>|z|) |
|
Intercept |
1,66 |
0,65 |
2,56 |
0,01030 |
|
Native American |
-2,80 |
0,75 |
-3,71 |
0,000202 |
|
European |
-0,10 |
0,88 |
-0,11 |
0,905630 |
|
African |
0,66 |
1,97 |
0,33 |
0,735190 |
In the model combining the three ancestries, it was found that only Native American genetic ancestry had a statistically significant effect (P < 0,001). Figure 3 shows the effect of Native American ancestry on the probability of carrying risk genotypes according to the logistic model reported here.
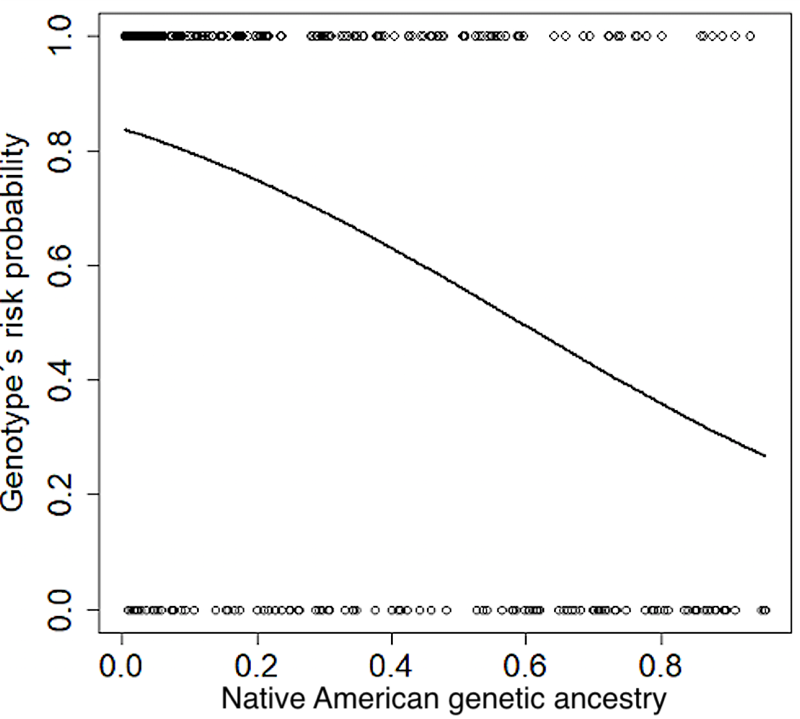
Figure 3. Predicted likelihood of VDR rs7975232 genotype`s risk for Native American genetic ancestry, from to the logistic regression model
DISCUSSION
The results of this study provide valuable insight into the distribution of rs7975232 (ApaI) genotypes in Latin American populations and their relationship with different ancestries. The absence of Hardy-Weinberg disequilibrium suggests that the sampled populations are genetically stable for this polymorphism, allowing for a reliable interpretation of the data.
The low proportion of Native American ancestry in the risk genotypes AA and CA is notable, indicating a possible lesser influence of this ancestry on the genetic predisposition to these genotypes. This finding contrasts with previous studies suggesting that certain ancestries may significantly impact genetic predisposition to diseases related to vitamin D deficiency and obesity.(7,9) The higher proportion of European ancestry in these risk genotypes reinforces the hypothesis that European genetic variability could contribute to a higher risk of developing conditions associated with vitamin D deficiency and increased BMI.(8,10)
The inverse relationship observed between Native American ancestry and the risk genotypes, as well as the positive relationship with European ancestry, suggests that genetic differences between these populations could influence the expression of the VDR gene and, therefore, vitamin D levels. This is consistent with studies indicating that variability in the VDR gene is associated with differences in vitamin D levels and the risk of metabolic diseases.(9)
Post hoc analyses with the Mann-Whitney test confirmed that the differences in the median ancestry between genotypes are significant, underscoring the importance of considering ancestries in genetic and epidemiological studies. Logistic regression showed that Native American ancestry has a significant inverse effect on the likelihood of carrying the risk genotypes, which could have important implications for public health and disease prevention in populations with a high proportion of Native American ancestry.
CONCLUSIONS
This study provides evidence on the influence of ancestries on the distribution of risk genotypes of the rs7975232 (ApaI) polymorphism in Latin American populations. The findings highlight the need for further research to better understand how genetic variability and ancestry interact to affect health and disease risk in these populations.
REFERENCES
1. Abdelaal M, le Roux CW, Docherty NG. Morbidity and mortality associated with obesity. Ann Transl Med. 2017;5(7):161–161. https://dx.doi.org/10.21037/atm.2017.03.107
2. Al-Rubeaan K, Bawazeer N, Al Farsi Y, Youssef AM, Al-Yahya AA, AlQumaidi H, et al. Prevalence of metabolic syndrome in Saudi Arabia - a cross sectional study. BMC Endocr Disord. 2018;18(1). https://dx.doi.org/10.1186/s12902-018-0244-4
3. Wortsman J, Matsuoka LY, Chen TC, Lu Z, Holick MF. Decreased bioavailability of vitamin D in obesity. Am J Clin Nutr. 2000;72(3):690–3. https://dx.doi.org/10.1093/ajcn/72.3.690
4. Liel Y, Shany S, Smirnoff P, Schwartz B. Estrogen Increases 1,25-Dihydroxyvitamin D Receptors Expression and Bioresponse in the Rat Duodenal Mucosa. Endocrinology. 1999;140(1):280–5. https://dx.doi.org/10.1210/endo.140.1.6408
5. Saminan S, Gusti N, Amirah S, Iqhrammullah M. Role of polymorphism in vitamin D receptor gene on diabetic foot ulcer: A systematic review and meta-analysis. J Pharm Pharmacogn Res . 2024;12(2):348–62. https://dx.doi.org/10.56499/jppres23.1790_12.2.348
6. Özhan B, Bi̇lgi̇han E, Çeti̇n O, Ağladioğlu K. The relations of vitamin D receptor gene polymorphisms with risk of obesity, metabolic syndrome, hepatostetosis in Turkish children. Pamukkale Med J. 2023;14–14. https://dx.doi.org/10.31362/patd.1249471
7. Rashidi F, Ostadsharif M. Association of VDR gene ApaI polymorphism with obesity in Iranian population. Biomedica. 2021;41(4):651–9. https://dx.doi.org/10.7705/biomedica.5898
8. Goh XX, Tee SF, Tang PY, Chee KY, Loh KKW. Impact of body mass index elevation, Vitamin D receptor polymorphisms and antipsychotics on the risk of Vitamin D deficiency in schizophrenia patients. J Psychiatr Res. 2024;175:350–8. https://dx.doi.org/10.1016/j.jpsychires.2024.05.005
9. Hassan NE, El-Masry SA, Zarouk WA, Eldeen GN, Mosaad RM, Afify MAS, et al. Narrative role of vitamin D receptor with osteoporosis and obesity in a sample of Egyptian females: a pilot study. J Genet Eng Biotechnol. 2021;19(1):115. https://dx.doi.org/10.1186/s43141-021-00216-0
10. Karuwanarint P, Phonrat B, Tungtrongchitr A, Suriyaprom K, Chuengsamarn S, Tungtronchitr R. Genetic variations of vitamin D receptor gene in metabolic syndrome and related diseases in the Thai population. Asia Pac J Clin Nutr . 2018;27(4):935–44. https://dx.doi.org/10.6133/apjcn.122017.04
11. Galanter JM, Fernandez-Lopez JC, Gignoux CR, Barnholtz-Sloan J, Fernandez-Rozadilla C, Via M, et al. Development of a panel of genome-wide ancestry informative markers to study admixture throughout the Americas. PLoS Genet. 2012;8(3). https://doi.org/10.1371/journal.pgen.1002554
12. Hubisz MJ, Falush D, Stephens M, Pritchard JK. Inferring weak population structure with the assistance of sample group information. Mol Ecol Resour. 2009;9(5):1322-1332. https://doi.org/10.1111/j.1755-0998.2009.02591.x
FINANCING
This research was funded by the “Regular Research Project Competition”, grant number “DI-04/23”, entitled “Prevalence of rs1861868-FTO and rs7975232-VDR polymorphisms in Chilean women and their association with BMI, anthropometry and cardiovascular risk factors. according to the consumption of estrogen-based contraceptives”, awarded by the Research Department of the University of the Americas, Providencia, Chile.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORSHIP CONTRIBUTION
Conceptualization: Sergio V. Flores.
Data curation: Sergio V. Flores, Román Montaña.
Formal analysis: Sergio V. Flores, Angel Roco-Videla.
Research: Sergio V. Flores, Angel Roco, Román Montaña, Marcela Caviedes-Olmos.
Methodology: Sergio V. Flores, Angel Roco-Videla, Sofia Pérez-Jiménez.
Software: Sergio V. Flores, Raúl Aguilera Eguía.
Supervision: Sergio V. Flores, Sofia Pérez-Jiménez.
Validation: Angel Roco-Videla, Marcela Caviedes-Olmos, Raúl Aguilera Eguía.
Display: Román Montaña, Sofia Pérez-Jiménez.
Drafting - original draft: Sergio V. Flores, Angel Roco-Videla.
Writing - proofreading and editing: Raúl Aguilera Eguía, Marcela Caviedes-Olmos.